<I>Neocosmospora</I>
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A New Species of Neocosmospora from Brazil
ZOBODAT - www.zobodat.at Zoologisch-Botanische Datenbank/Zoological-Botanical Database Digitale Literatur/Digital Literature Zeitschrift/Journal: Sydowia Jahr/Year: 1995 Band/Volume: 47 Autor(en)/Author(s): Pfenning Ludwig Artikel/Article: A new species of Neocosmospora from Brazil. 65-69 ©Verlag Ferdinand Berger & Söhne Ges.m.b.H., Horn, Austria, download unter www.biologiezentrum.at A new species of Neocosmospora from Brazil Ludwig Pfenning* Lehrstuhl Spezielle Botanik/Mykologie, Universität Tübingen, Auf der Morgenstelle 1, D- 72076 Tübingen, Germany Pfenning, L. (1995). A new species of Neocosmospora from Brazil. Sydowia 47 (1): 65-69. During an investigation of rhizosphere fungi of primary forests and cultivated areas in the Brazilian Amazon a new species of Neocosmospora (Hypocreales, Ascomycetes) with spinose ascospores was isolated. The name N. spinulosa is proposed. Keywords: anamorph-teleomorph connection, rhizosphere, taxonomy. The genus Neocosmospora was established by E. F. Smith in 1899, with the type species N. vasinfecta. The genus, classified in the order Hypocreales (Ascomycetes) fam. Hypocreaceae, is characterized by the yellowish to reddish-brown, membranous wall of the ascomata, asci without apical differentiations and thick-walled, ornamented ascospores lacking germ pores. The species have mostly been isolated from soil or roots in tropical and subtropical regions. Most of the known anamorphs are Acremonium spp., but that of N. endophytica Polishook & al. (Polishook & al., 1991) belongs to Penicillifer van Emden. The genus was revised by Cannon & Hawksworth (1984) who accepted five species and one variety, synonymizing N. ornamentata Freitas Barbosa (Freitas Barbosa, 1965) with N. vasinfecta E. F. Sm. var. vasinfecta. Two new species from Japan, N. -

New Insights Into the Microbiota of Moth Pests
International Journal of Molecular Sciences Review New Insights into the Microbiota of Moth Pests Valeria Mereghetti, Bessem Chouaia and Matteo Montagna * ID Dipartimento di Scienze Agrarie e Ambientali, Università degli Studi di Milano, 20122 Milan, Italy; [email protected] (V.M.); [email protected] (B.C.) * Correspondence: [email protected]; Tel.: +39-02-5031-6782 Received: 31 August 2017; Accepted: 14 November 2017; Published: 18 November 2017 Abstract: In recent years, next generation sequencing (NGS) technologies have helped to improve our understanding of the bacterial communities associated with insects, shedding light on their wide taxonomic and functional diversity. To date, little is known about the microbiota of lepidopterans, which includes some of the most damaging agricultural and forest pests worldwide. Studying their microbiota could help us better understand their ecology and offer insights into developing new pest control strategies. In this paper, we review the literature pertaining to the microbiota of lepidopterans with a focus on pests, and highlight potential recurrent patterns regarding microbiota structure and composition. Keywords: symbiosis; bacterial communities; crop pests; forest pests; Lepidoptera; next generation sequencing (NGS) technologies; diet; developmental stages 1. Introduction Insects represent the most successful taxa of eukaryotic life, being able to colonize almost all environments, including Antarctica, which is populated by some species of chironomids (e.g., Belgica antarctica, Eretmoptera murphyi, and Parochlus steinenii)[1,2]. Many insects are beneficial to plants, playing important roles in seed dispersal, pollination, and plant defense (by feeding upon herbivores, for example) [3]. On the other hand, there are also damaging insects that feed on crops, forest and ornamental plants, or stored products, and, for these reasons, are they considered pests. -

Calonectria Ilicicola
OctoberPathogen of11 the month –October 2011 a b c d Reitsma & Fig. 1. Calonectria ilicicola; asci with ascospores (a); microsclerotia on a papaya root (b); conidium of anamorph, Cylindrocladium parasiticum (c); orange perithecia on papaya root (d); Photo credits M. Male (a,b,c), L. Vawdrey (d) Disease: Collar rot of papaya Pathogen: Calonectria ilicicola (anamorph: Cylindrocladium parasiticum) Boedijn Classification: K: Fungi, D: Ascomycota, C: Sordariomycetes, O: Hypocreales, F: Nectriaceae Calonectria ilicicola (Fig. 1) is a fungal pathogen found throughout the world infecting a wide variety of crops. It is best known as the causal agent of peg, pod and root necrosis of peanuts, collar rot of koa, leaf spot in some eucalypts and various root and collar rots of soybean, anthurium, groundnut and lucerne. In Australia, it causes black rot of peanut and collar rot of papaya. Its development is favoured by poorly drained soils. Host Range: Key Distinguishing Features: C. ilicicola is found throughout the world. It is In seedlings, the initial symptoms are of a discoloured pathogenic on a wide variety of plants including water soaked, root collar. As this rot develops, plants peanuts, soybean, anthurium, eucalypts, lucerne, are stunted and leaves become chlorotic and wilt. groundnut and papaya. Eventually, orange to red perithecia and thick-walled ilicicola brown microsclerotia will form on roots at or near the Impact: soil line (Fig 1 b,d). In culture, asci are clavate, 90- In far north Queensland, collar rot of papaya caused 140 x 12-19μm, tapering to a long thin stalk by C. ilicicola affects young plants in poorly drained containing eight hyaline ascospores (Fig1,a). -

Illuminating Type Collections of Nectriaceous Fungi in Saccardo's
Persoonia 45, 2020: 221–249 ISSN (Online) 1878-9080 www.ingentaconnect.com/content/nhn/pimj RESEARCH ARTICLE https://doi.org/10.3767/persoonia.2020.45.09 Illuminating type collections of nectriaceous fungi in Saccardo’s fungarium N. Forin1, A. Vizzini 2,3,*, S. Nigris1,4, E. Ercole2, S. Voyron2,3, M. Girlanda2,3, B. Baldan1,4,* Key words Abstract Specimens of Nectria spp. and Nectriella rufofusca were obtained from the fungarium of Pier Andrea Saccardo, and investigated via a morphological and molecular approach based on MiSeq technology. ITS1 and ancient DNA ITS2 sequences were successfully obtained from 24 specimens identified as ‘Nectria’ sensu Saccardo (including Ascomycota 20 types) and from the type specimen of Nectriella rufofusca. For Nectria ambigua, N. radians and N. tjibodensis Hypocreales only the ITS1 sequence was recovered. On the basis of morphological and molecular analyses new nomenclatural Illumina combinations for Nectria albofimbriata, N. ambigua, N. ambigua var. pallens, N. granuligera, N. peziza subsp. ribosomal sequences reyesiana, N. radians, N. squamuligera, N. tjibodensis and new synonymies for N. congesta, N. flageoletiana, Sordariomycetes N. phyllostachydis, N. sordescens and N. tjibodensis var. crebrior are proposed. Furthermore, the current classifi- cation is confirmed for Nectria coronata, N. cyanostoma, N. dolichospora, N. illudens, N. leucotricha, N. mantuana, N. raripila and Nectriella rufofusca. This is the first time that these more than 100-yr-old specimens are subjected to molecular analysis, thereby providing important new DNA sequence data authentic for these names. Article info Received: 25 June 2020; Accepted: 21 September 2020; Published: 23 November 2020. INTRODUCTION to orange or brown perithecia which do not change colour in 3 % potassium hydroxide (KOH) or 100 % lactic acid (LA) Nectria, typified with N. -

Cylindrocladium Buxicola Nom. Cons. Prop.(Syn. Calonectria
I Promotors: Prof. dr. ir. Monica Höfte Laboratory of Phytopathology, Department of Crop Protection Faculty of Bioscience Engineering Ghent University Dr. ir. Kurt Heungens Institute for Agricultural and Fisheries Research (ILVO) Plant Sciences Unit - Crop Protection Dean: Prof. dr. ir. Guido Van Huylenbroeck Rector: Prof. dr. Anne De Paepe II Bjorn Gehesquière Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) on Buxus: molecular characterization, epidemiology, host resistance and fungicide control Thesis submitted in fulfillment of the requirements for the degree of Doctor (PhD) in Applied Biological Sciences III Dutch translation of the title: Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) in Buxus: moleculaire karakterisering, epidemiologie, waardplantresistentie en chemische bestrijding. Please refer to this work as follows: Gehesquière B. (2014). Cylindrocladium buxicola nom. cons. prop. (syn. Calonectria pseudonaviculata) on Buxus: molecular characterization, epidemiology, host resistance and fungicide control. Phd Thesis. Ghent University, Belgium The author and the promotors give authorisation to consult and to copy parts of this work for personal use only. Any other use is limited by Laws of Copyright. Permission to reproduce any material contained in this work should be obtained from the author. The promotors, The author, Prof. dr. ir. M. Höfte Dr. ir. K. Heungens ir. B. Gehesquière IV Een woordje van dank…. Dit dankwoord schrijven is ongetwijfeld het leukste onderdeel van deze thesis, en een mooie afsluiting van een interessante periode. Terugblikkend op de voorbije vier jaren kan ik enkel maar beamen dat een doctoraat zoveel meer is dan een wetenschappelijke uitdaging. Het is een levensreis in al zijn facetten, waarbij ik mezelf heb leren kennen in al mijn goede en slechte kantjes. -

Epidemiological Studies on the Infection Process and Symptom Expression of Soybean Sudden Death Syndrome Carlos Cecilio Gongora-Canul Iowa State University
Iowa State University Capstones, Theses and Graduate Theses and Dissertations Dissertations 2010 Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome Carlos Cecilio Gongora-canul Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/etd Part of the Plant Pathology Commons Recommended Citation Gongora-canul, Carlos Cecilio, "Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome" (2010). Graduate Theses and Dissertations. 11510. https://lib.dr.iastate.edu/etd/11510 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. Epidemiological studies on the infection process and symptom expression of soybean sudden death syndrome by Carlos Cecilio Góngora-Canul A dissertation submitted to the graduate faculty in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Major: Plant Pathology Program of Study Committee: Leonor Leandro, Major Professor Gary Munkvold Greg Tylka X. B Yang Dan Nordman Iowa State University Ames, Iowa 2010 Copyright © Carlos Cecilio Góngora-Canul, 2010. All rights reserved. ii DEDICATION To my Lord, for giving me the blessing and the adventure to live. To my mother Elsa and my father Elias for their endless love and to all my brothers and sisters (Javier, Roberto, Martha, Manuel, Enrique and Nicte-Há) for all their great love affection. -
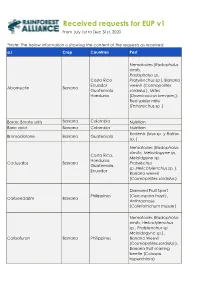
IIS 2020 Requests Overview
Received requests for EUP v1 From July 1st to Dec 31st, 2020 *Note: The below information is showing the content of the requests as received. a.i Crop Countries Pest Nematodes ( Radophulus similis, Radopholus sp, Costa Rica Pratylenchus sp ), Banana Ecuador weevil ( Cosmopolites Abamectin Banana Guatemala sordidus ), Mites Honduras (Dysmicoccus brevipes ), Red spider mite (Tetranychus sp .) Borax; Borate salts Banana Colombia Nutrition Boric acid Banana Colombia Nutrition Rodents ( Mus sp. y Rattus Bromadiolone Banana Guatemala sp. ) Nematodes ( Radopholus silmillis, Meloidogyne sp, Costa Rica, Meloidgune sp, Honduras, Cadusafos Banana Pratylechus Guatemala, sp.,Helicotylenchus sp. ), Ecuador Banana weevil (Cosmopolites sordidus ) Diamond Fruit Sport Philippines (Cercospora hayii ), Carbendazim Banana Anthracnose (Colletotrichum musae ) Nematod es (Radopholus similis, Helicotylenchus sp., Pratylenchus sp. Meloidogyne sp.) , Carbofuran Banana Philippines Banana Weevil (Cosmopolites sordidus ), Banana fruit scarring beetle (Colaspis hyperchlora) a.i Crop Countries Pest Black Sigatoka Ecuador, Costa (Mycosphaerella fijiensis ), Rica, Philippines, Yellow Sigatoka Chlorothalonil Banana Honduras, (Mycosphaerella Guatemala, musicola ), Banana Colombia Freckle ( Phyllosticta musarum ) Mealybug ( Dysmicoccus brevipes, Pseudococcus elisae, Pseudococcus sp, Ferrista sp ), Dysmicoccus sp , Scale insects (Aspidiotus destructor, Diaspis boisduvalii ), Thrips (Thrips florum, Franckliniella spp, Chaetanaphothrips signipennis ), Banana fruit Philippines, -

Pathogenicity and Molecular Detection of Nectriaceous Fungi Associated with Black Root Rot of Avocado
IX World Avocado Congress, 23 – 27 September, 2019, Medellín, Colombia WAC-130 PATHOGENICITY AND MOLECULAR DETECTION OF NECTRIACEOUS FUNGI ASSOCIATED WITH BLACK ROOT ROT OF AVOCADO L. E. Parkinson, D. P. Le, R. G. Shivas and E. K. Dann Dr Louisamarie Parkinson BBiotech(Hons), PhD Centre for Horticultural Science Queensland Alliance for Agriculture and Food Innovation (QAAFI) The University of Queensland, Brisbane Qld 4072 Australia [email protected] | https://www.qaafi.uq.edu.au THE UNIVERSITY OF QUEENSLAND St Lucia, Brisbane, Queensland Australia PATHOGENICITY AND MOLECULAR DETECTION OF NECTRIACEOUS FUNGI ASSOCIATED WITH BLACK ROOT ROT OF AVOCADO L. E. Parkinson1, D. P. Le1, R. G. Shivas2, E. K. Dann1 1 Queensland Alliance for Agriculture and Food Innovation, The University of Queensland, Australia 2Centre for Crop Health, The University of Southern Queensland, Australia KEY WORDS Calonectria, Calonectria ilicicola, Dactylonectria, Dactylonectria macrodidyma, diagnostic test, diversity, loop-mediated isothermal amplification (LAMP) SUMMARY Black root rot of avocado associated with soilborne nectriaceous fungi is an aggressive disease of nursery trees and young orchards transplants, causing tree stunting, wilt, severe root necrosis, rapid decline and death within a year after planting. This study aimed to identify the fungal genera associated with the disease, determine the causal agents of black root rot, and develop a rapid molecular test for detection of key pathogens in avocado roots. A disease survey in all Australian growing regions collected 153 nectriaceous fungal isolates from roots of 91 symptomatic and healthy avocado trees and other hosts including peanut, papaya, blueberry, custard apple and grapevine. The fungal isolates were identified with phylogenetic analyses of ITS, β-tubulin and Histone H3 sequenced genes. -
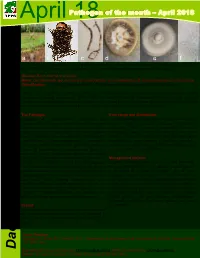
Dactylonectria Lombard & Crous
April Pathogen18 of the month – April 2018 a b c d e f Fig. 1. Black root rot symptoms in young orchard transplants (a), necrotic avocado roots (b, c) Dactylonectria macrodidyma on ½ sPDA at 3.75 cm after 10 days growth (d, e), D. macrodidyma macroconidia at 40 × magnification (f) Disease: Black root rot of avocado Name: Dactylonectria spp. including D. macrodidyma, D. novozelandica, D. pauciseptata and D. anthuriicola Classification: K: Fungi, D: Ascomycota, C: Sordariomycetes, O: Hypocreales, F: Nectriaceae Black root rot caused by nectriaceous fungi is a severe disease of avocado nursery trees and young orchard transplants, causing decline and death within one year of planting. Symptoms include stunting, wilt, leaf chlorosis and browning, leaf drop prior to tree death caused by severe necrosis of the root system. In Australia black root rot of avocado is caused by Calonectria ilicicola and several Dactylonectria spp. The Pathogen: Host range and distribution: Species of Dactylonectria (reported as Dactylonectria spp. cause root rot diseases in various Cylindrocarpon in older literature) have often been hosts including avocado (Persea americana), isolated from necrotic avocado roots. Dactylonectria grapevine (Vitis vinifera), cherimoya (Annona macrodidyma is the most prevalent of the pathogens cherimola), kiwifruit (Actinidia deliciosa) and olive found in symptomatic avocado roots. Dactylonectria (Olea europaea). Dactylonectria spp. associated with novozelandica, D. pauciseptata and D. anthuriicola avocado have been reported in Australia and Italy. have also been isolated from avocado roots and However the fungal genus is reported globally across Lombard & Crous shown to be pathogenic in glasshouse tests with numerous horticultural industries. seedlings. While Dactylonectria spp. -

(Hypocreales) Proposed for Acceptance Or Rejection
IMA FUNGUS · VOLUME 4 · no 1: 41–51 doi:10.5598/imafungus.2013.04.01.05 Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) ARTICLE proposed for acceptance or rejection Amy Y. Rossman1, Keith A. Seifert2, Gary J. Samuels3, Andrew M. Minnis4, Hans-Josef Schroers5, Lorenzo Lombard6, Pedro W. Crous6, Kadri Põldmaa7, Paul F. Cannon8, Richard C. Summerbell9, David M. Geiser10, Wen-ying Zhuang11, Yuuri Hirooka12, Cesar Herrera13, Catalina Salgado-Salazar13, and Priscila Chaverri13 1Systematic Mycology & Microbiology Laboratory, USDA-ARS, Beltsville, Maryland 20705, USA; corresponding author e-mail: Amy.Rossman@ ars.usda.gov 2Biodiversity (Mycology), Eastern Cereal and Oilseed Research Centre, Agriculture & Agri-Food Canada, Ottawa, ON K1A 0C6, Canada 3321 Hedgehog Mt. Rd., Deering, NH 03244, USA 4Center for Forest Mycology Research, Northern Research Station, USDA-U.S. Forest Service, One Gifford Pincheot Dr., Madison, WI 53726, USA 5Agricultural Institute of Slovenia, Hacquetova 17, 1000 Ljubljana, Slovenia 6CBS-KNAW Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands 7Institute of Ecology and Earth Sciences and Natural History Museum, University of Tartu, Vanemuise 46, 51014 Tartu, Estonia 8Jodrell Laboratory, Royal Botanic Gardens, Kew, Surrey TW9 3AB, UK 9Sporometrics, Inc., 219 Dufferin Street, Suite 20C, Toronto, Ontario, Canada M6K 1Y9 10Department of Plant Pathology and Environmental Microbiology, 121 Buckhout Laboratory, The Pennsylvania State University, University Park, PA 16802 USA 11State -

Color Plates
Color Plates Plate 1 (a) Lethal Yellowing on Coconut Palm caused by a Phytoplasma Pathogen. (b, c) Tulip Break on Tulip caused by Lily Latent Mosaic Virus. (d, e) Ringspot on Vanda Orchid caused by Vanda Ringspot Virus R.K. Horst, Westcott’s Plant Disease Handbook, DOI 10.1007/978-94-007-2141-8, 701 # Springer Science+Business Media Dordrecht 2013 702 Color Plates Plate 2 (a, b) Rust on Rose caused by Phragmidium mucronatum.(c) Cedar-Apple Rust on Apple caused by Gymnosporangium juniperi-virginianae Color Plates 703 Plate 3 (a) Cedar-Apple Rust on Cedar caused by Gymnosporangium juniperi.(b) Stunt on Chrysanthemum caused by Chrysanthemum Stunt Viroid. Var. Dark Pink Orchid Queen 704 Color Plates Plate 4 (a) Green Flowers on Chrysanthemum caused by Aster Yellows Phytoplasma. (b) Phyllody on Hydrangea caused by a Phytoplasma Pathogen Color Plates 705 Plate 5 (a, b) Mosaic on Rose caused by Prunus Necrotic Ringspot Virus. (c) Foliar Symptoms on Chrysanthemum (Variety Bonnie Jean) caused by (clockwise from upper left) Chrysanthemum Chlorotic Mottle Viroid, Healthy Leaf, Potato Spindle Tuber Viroid, Chrysanthemum Stunt Viroid, and Potato Spindle Tuber Viroid (Mild Strain) 706 Color Plates Plate 6 (a) Bacterial Leaf Rot on Dieffenbachia caused by Erwinia chrysanthemi.(b) Bacterial Leaf Rot on Philodendron caused by Erwinia chrysanthemi Color Plates 707 Plate 7 (a) Common Leafspot on Boston Ivy caused by Guignardia bidwellii.(b) Crown Gall on Chrysanthemum caused by Agrobacterium tumefaciens 708 Color Plates Plate 8 (a) Ringspot on Tomato Fruit caused by Cucumber Mosaic Virus. (b, c) Powdery Mildew on Rose caused by Podosphaera pannosa Color Plates 709 Plate 9 (a) Late Blight on Potato caused by Phytophthora infestans.(b) Powdery Mildew on Begonia caused by Erysiphe cichoracearum.(c) Mosaic on Squash caused by Cucumber Mosaic Virus 710 Color Plates Plate 10 (a) Dollar Spot on Turf caused by Sclerotinia homeocarpa.(b) Copper Injury on Rose caused by sprays containing Copper. -

The Effects of Climate Change on the Pests and Diseases of Coffee
y & W log ea to th a e im r l F o Journal of Climatology & Weather C f r e o c l a a s n Groenen, Climatol Weather Forecasting 2018, 6:3 t r i n u g o J Forecasting DOI: 10.4172/2332-2594.1000239 ISSN: 2332-2594 Review Article Open Access The Effects of Climate Change on the Pests and Diseases of Coffee Crops in Mesoamerica Danielle Groenen* Department of Earth, Ocean and Atmospheric Science and Center for Ocean–Atmospheric Prediction Studies, Florida State University, Florida, United States *Corresponding author: Danielle Groenen, Department of Earth, Ocean and Atmospheric Science and Center for Ocean–Atmospheric Prediction Studies, Florida State University, Florida, United States, Tel: 18506442525; E-mail: [email protected] Received Date: August 30, 2018; Accepted Date: September 21, 2018; Published Date: September 26, 2018 Copyright: © 2018 Groenen D. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. Abstract Coffee is an in-demand commodity that is being threatened by climate change. Increasing temperatures and rainfall variability are predicted in the region of Mexico and Central America (Mesoamerica). This region is plagued with pests and diseases that have already caused millions of dollars in damages and losses to the coffee industry. This paper examines three pests that negatively affect coffee plants: the coffee borer beetle, the black twig borer, and nematodes. In addition, this paper examines three diseases that can destroy coffee crops: bacterial blight, coffee berry disease, and coffee leaf rust.