Server-Aided 3D DICOM Viewer for Mobile Platforms
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -

Survey of Databases Used in Image Processing and Their Applications
International Journal of Scientific & Engineering Research Volume 2, Issue 10, Oct-2011 1 ISSN 2229-5518 Survey of Databases Used in Image Processing and Their Applications Shubhpreet Kaur, Gagandeep Jindal Abstract- This paper gives review of Medical image database (MIDB) systems which have been developed in the past few years for research for medical fraternity and students. In this paper, I have surveyed all available medical image databases relevant for research and their use. Keywords: Image database, Medical Image Database System. —————————— —————————— 1. INTRODUCTION Measurement and recording techniques, such as electroencephalography, magnetoencephalography Medical imaging is the technique and process used to (MEG), Electrocardiography (EKG) and others, can create images of the human for clinical purposes be seen as forms of medical imaging. Image Analysis (medical procedures seeking to reveal, diagnose or is done to ensure database consistency and reliable examine disease) or medical science. As a discipline, image processing. it is part of biological imaging and incorporates radiology, nuclear medicine, investigative Open source software for medical image analysis radiological sciences, endoscopy, (medical) Several open source software packages are available thermography, medical photography and for performing analysis of medical images: microscopy. ImageJ 3D Slicer ITK Shubhpreet Kaur is currently pursuing masters degree OsiriX program in Computer Science and engineering in GemIdent Chandigarh Engineering College, Mohali, India. E-mail: MicroDicom [email protected] FreeSurfer Gagandeep Jindal is currently assistant processor in 1.1 Images used in Medical Research department Computer Science and Engineering in Here is the description of various modalities that are Chandigarh Engineering College, Mohali, India. E-mail: used for the purpose of research by medical and [email protected] engineering students as well as doctors. -
PDF File, 212 KB
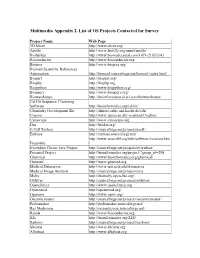
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -

Whole Body Computed Tomography with Advanced Imaging Techniques: a Research Tool for Measuring Body Composition in Dogs
Hindawi Publishing Corporation Journal of Veterinary Medicine Volume 2013, Article ID 610654, 6 pages http://dx.doi.org/10.1155/2013/610654 Research Article Whole Body Computed Tomography with Advanced Imaging Techniques: A Research Tool for Measuring Body Composition in Dogs Dharma Purushothaman,1 Barbara A. Vanselow,2 Shu-Biao Wu,1 Sarah Butler,3 and Wendy Yvonne Brown1 1 School of Environmental and Rural Science, Department of Animal Science, University of New England, Armidale, NSW 2351, Australia 2 NSW Department of Primary Industries, Beef Industry Centre, University of New England, Armidale, NSW 2351, Australia 3 North Hill Vet Clinic, Armidale, NSW 2350, Australia Correspondence should be addressed to Wendy Yvonne Brown; [email protected] Received 6 May 2013; Revised 14 September 2013; Accepted 17 September 2013 Academic Editor: Juan G. Chediack Copyright © 2013 Dharma Purushothaman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The use of computed tomography (CT) to evaluate obesity in canines is limited. Traditional CT image analysis is cumbersome and uses prediction equations that require manual calculations. In order to overcome this, our study investigated the use of advanced image analysis software programs to determine body composition in dogs with an application to canine obesity research. Beagles and greyhounds were chosen for their differences in morphology and propensity to obesity. Whole body CT scans with regular intervals were performed on six beagles and six greyhounds that were subjected to a 28-day weight-gain protocol. -

Evaluation of DICOM Viewer Software for Workflow Integration in Clinical Trials
Evaluation of DICOM Viewer Software for Workflow Integration in Clinical Trials Daniel Haak1*, Charles-E. Page, Klaus Kabino, Thomas M. Deserno Department of Medical Informatics, Uniklinik RWTH Aachen, 52057 Aachen, Germany ABSTRACT The digital imaging and communications in medicine (DICOM) protocol is nowadays the leading standard for capture, exchange and storage of image data in medical applications. A broad range of commercial, free, and open source software tools supporting a variety of DICOM functionality exists. However, different from patient’s care in hospital, DICOM has not yet arrived in electronic data capture systems (EDCS) for clinical trials. Due to missing integration, even just the visualization of patient’s image data in electronic case report forms (eCRFs) is impossible. Four increasing levels for integration of DICOM components into EDCS are conceivable, raising functionality but also demands on interfaces with each level. Hence, in this paper, a comprehensive evaluation of 27 DICOM viewer software projects is performed, investigating viewing functionality as well as interfaces for integration. Concerning general, integration, and viewing requirements the survey involves the criteria (i) license, (ii) support, (iii) platform, (iv) interfaces, (v) two- dimensional (2D) and (vi) three-dimensional (3D) image viewing functionality. Optimal viewers are suggested for applications in clinical trials for 3D imaging, hospital communication, and workflow. Focusing on open source solutions, the viewers ImageJ and MicroView are superior for 3D visualization, whereas GingkoCADx is advantageous for hospital integration. Concerning workflow optimization in multi-centered clinical trials, we suggest the open source viewer Weasis. Covering most use cases, an EDCS and PACS interconnection with Weasis is suggested. -

Kitware Source Issue 21
SOFTWARE DEVELOPER’S QUARTERLY Issue 21 • April 2012 Editor’s Note ........................................................................... 1 PARAVIEW 3.14 RELEASED In late February, Kitware and the ParaView team released Recent Releases ..................................................................... 1 ParaView 3.14. This release features usability enhancements, improvements to the Plugin framework, new panels, and ParaView in Immersive Environments .................................. 3 more than 100 other resolved issues. Teaching Open Source .......................................................... 4 ParaView 3.14 features a redesigned Find Data dialog and Color Editor. The updated Find Data dialog makes it possible ParaView Query Selection Framework................................. 7 to use complex queries to select elements, including combin- ing multiple test cases with Boolean operations. The Color Ginkgo CADx: Open Source DICOM CADx Environment .... 8 Editor, used to edit lookup tables or color tables for scalar mapping, now enables independent editing of the color and Code Review, Topic Branches and VTK ................................. 9 opacity functions. Video Analysis on Business Intelligence, ParaView’s charting capabilities have been extended with a Studies in Computational Intelligence ............................... 11 new scatter plot matrix view and the ability to visualize mul- tiple dimensions of data in one compact form. This improved The Visible Patient .............................................................. -

Transforming the Medical Imaging Workflow
Transforming the Medical Imaging Workflow: How Mac systems and open source software combine to make full-featured diagnostic imaging solutions affordable for today’s radiologist. By Roger Katen, M.D. San Francisco, CA White Paper 2 Transforming the Medical Imaging Workflow Contents Page 3 Executive Summary Page 5 The New Realities of Medical Imaging The Digital Data Explosion The Rise of Imaging Workstations A Cost Conundrum Page 8 A Compelling Alternative: The Mac and OsiriX Workstation OsiriX: Full-Featured, Open Source Mac OS X and Mac Systems A New Solution for a New Reality Page 10 Mac Systems and OsiriX in the Radiology Workflow Image Generation Equipment Picture Archiving and Communications Systems (PACS) Creating a PACS Archive with OsiriX Database Sharing and Autorouting Storage System Page 16 Building an Imaging Workstation Environment with OsiriX Working with Large Datasets Supporting Medical-Quality Displays Requirements for High-Performance Processors Accelerating Image Retrieval Page 19 Enabling the Imaging Workflow Options for Advanced Visualization Starting and Sending a New DICOM Series to PACS Emailing and Printing Images Integrating with Hospital and Radiology Information Systems Exploring Alternatives to Windows RIS Systems Page 24 Collaborating Beyond the Office with OsiriX Virtual Private Networks Remote Visualization via Apple Remote Desktop Citrix/Windows Terminal Services Teleradiology Collaboration iChat and iChat Theater Page 28 Conclusion White Paper 3 Transforming the Medical Imaging Workflow Executive Summary Today more than ever, the healthcare industry is reaping the benefits of advances in diagnostic medical imaging. Dramatic breakthroughs in higher-resolution CT, MRI, ultrasound, and interventional technologies have enabled healthcare providers to deliver more informed diagnoses, pursue more effective treatments, collaborate more easily with colleagues, and communicate more clearly with patients. -

Automated 3D Visualization of Brain Cancer
AUTOMATED 3D VISUALIZATION OF BRAIN CANCER AUTOMATED 3D VISUALIZATION OF BRAIN CANCER By MONA AL-REI, MSc. A Thesis Submitted to the School of Graduate Studies In Partial Fulfillment of the Requirements for the Degree Master of eHealth Program McMaster University @ Copyright by Mona Al-Rei, June 2017 McMaster University Master of eHealth (2017) Hamilton, Ontario TITLE: 3D Brain Cancer Visualization. AUTHOR: Mona Al-Rei. SUPERVISOR: Dr. Thomas Doyle. SUPERVISRORY COMMITTEE: Dr. Reza Samavi, Dr. David Koff. NUMBER OF PAGES: xvii, 119. ii To my beloved and wounded country Yemen iii Abstract Three-dimensional (3D) visualization in cancer control has seen recent progress due to the benefits it offers to the treatment, education, and understanding of the disease. This work identifies the need for an application that directly processes two-dimensional (2D) DICOM images for the segmentation of a brain tumor and the generation of an interactive 3D model suitable for enabling multisensory learning and visualization. A new software application (M-3Ds) was developed to meet these objectives with three modes of segmentation (manual, automatic, and hybrid) for evaluation. M-3Ds software was designed to mitigate the cognitive load and empower health care professionals in their decision making for improved patient outcomes and safety. Comparison of mode accuracy was evaluated. Industrial standard software programs were employed to verify and validate the results of M-3Ds using quantitative volumetric comparison. The study determined that M-3Ds‘ hybrid mode was the highest accuracy with least user intervention for brain tumor segmentation and suitable for the clinical workflow. This paper presents a novel approach to improve medical education, diagnosis, treatment for either surgical planning or radiotherapy of brain cancer. -

MRI and CT Fusion in Stereotactic Electroencephalography: a Literature Review
applied sciences Systematic Review MRI and CT Fusion in Stereotactic Electroencephalography: A Literature Review Jaime Perez 1,* , Claudia Mazo 1,2,3 , Maria Trujillo 1 and Alejandro Herrera 1,4 1 Multimedia and Computer Vision Group, Universidad del Valle, Cali 760001, Colombia; [email protected] (C.M.); [email protected] (M.T.); [email protected] (A.H.) 2 UCD School of Computer Science, University College Dublin, Dublin 4, Ireland 3 CeADAR Ireland’s Centre for Applied AI, Dublin 4, Ireland 4 Clinica Imbanaco Grupo Quironsalud, Cali 760001, Colombia * Correspondence: [email protected] Abstract: Epilepsy is a common neurological disease characterized by spontaneous recurrent seizures. Resection of the epileptogenic tissue may be needed in approximately 25% of all cases due to ineffec- tive treatment with anti-epileptic drugs. The surgical intervention depends on the correct detection of epileptogenic zones. The detection relies on invasive diagnostic techniques such as Stereotactic Electroencephalography (SEEG), which uses multi-modal fusion to aid localizing electrodes, using pre-surgical magnetic resonance and intra-surgical computer tomography as the input images. More- over, it is essential to know how to measure the performance of fusion methods in the presence of external objects, such as electrodes. In this paper, a literature review is presented, applying the methodology proposed by Kitchenham to determine the main techniques of multi-modal brain image fusion, the most relevant performance metrics, and the main fusion tools. The search was conducted using the databases and search engines of Scopus, IEEE, PubMed, Springer, and Google Citation: Perez, J.; Mazo, C.; Trujillo, Scholar, resulting in 15 primary source articles. -

Segmentation Tools Frederic Cervenansky What Is Open-Source
Through the forest of open source segmentation tools Frederic Cervenansky What is Open-Source Open source doesn’t just mean access to source code 1 - Free Redistribution (no restriction from selling or giving away the software as a component) 2 - Source Code ( as well as compiled form) 3 - Derived Works (copyleft license) 4 - Integrity of The Author's Source Code (users have a right to know who is the original author) 5 – Distribution of License (no need of a third license) 6 and more What criteria to discriminate software? Usability Code Langage Segmentation methods Plugins – Extensions Organ/structure specificity Software Organs specific Code Segmentation Plugins Citations* Language Methods ImageJ no Java Automatic Yes 723 Fiji No ( Cell) Java Automatic Yes 1254 Freesurfer Brain C++, bash Automatic no 470 ITK no C++ Automatic Yes-no 512 Slicer3D No (Brain) C++ Manual- Automatic Yes 75 FSL Brain C++, bash Automatic No 690 SPM Brain matlab Automatic Yes 7574 pubmed … --- --- --- --- --- *: JAVA Bio-imaging Plugins system User friendly IMAGEJ 2D and 3D limitation No hierarchy in plugins ImgLib (n-dimensional, repackaging, sharing improvement) KMeans Color Quantization Color Picker Fiji Marching IMAGEJ2 Icy Threshold Learning Active Contours Squassh KMeans Clustering Texture Level Set Maximum Entropy Threshold HK-Means Graph Cut Maximum Multi Entropy Threshold Active Cells Bad Indexation: Seeded Region Growing Toolbox dithering as Snake. segmenation tool Brain Organ specific Fully automatic FreeSurfer Allow to restart at divergent steps. Both available Fully automatic through VIP Catalog of tools FSL BET ( Brain Extraction) FAST (GM and WM automated Segmentation). SPM Modified gaussian mixture model Need a matlab license.