Neuroimaging DICOM and Nifti Primer
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

Modern Programming Languages CS508 Virtual University of Pakistan
Modern Programming Languages (CS508) VU Modern Programming Languages CS508 Virtual University of Pakistan Leaders in Education Technology 1 © Copyright Virtual University of Pakistan Modern Programming Languages (CS508) VU TABLE of CONTENTS Course Objectives...........................................................................................................................4 Introduction and Historical Background (Lecture 1-8)..............................................................5 Language Evaluation Criterion.....................................................................................................6 Language Evaluation Criterion...................................................................................................15 An Introduction to SNOBOL (Lecture 9-12).............................................................................32 Ada Programming Language: An Introduction (Lecture 13-17).............................................45 LISP Programming Language: An Introduction (Lecture 18-21)...........................................63 PROLOG - Programming in Logic (Lecture 22-26) .................................................................77 Java Programming Language (Lecture 27-30)..........................................................................92 C# Programming Language (Lecture 31-34) ...........................................................................111 PHP – Personal Home Page PHP: Hypertext Preprocessor (Lecture 35-37)........................129 Modern Programming Languages-JavaScript -

TANGO Device
School on TANGO Controls system Basics of TANGO Lorenzo Pivetta Claudio Scafuri Graziano Scalamera http://www.tango-controls.org L.Pivetta, C.Scafuri, G.Scalamera School on TANGO Control System - Trieste 4-8th July 2016 2 Prerequisites To better understand the training a background on the following arguments is desirable: ● Programming language ● Object oriented programming ● Linux/UNIX operating system ● Networking ● Control systems L.Pivetta, C.Scafuri, G.Scalamera School on TANGO Control System - Trieste 4-8th July 2016 3 Outline 1 - What is TANGO? 2 - TANGO architecture Language/OS/Compilers Device hierarchy CORBA and ZeroMQ TANGO domains TANGO device and device server TANGO Database Communication models 3 - TANGO configuration/tools Multicast Jive Polling Starter/Astor Events Pogo Alarms TANGO installation Groups Client basics TANGO ACL Logging system Historical DataBase 4 – Examples Test device L.Pivetta, C.Scafuri, G.Scalamera School on TANGO Control System - Trieste 4-8th July 2016 4 What is TANGO? Scientific workspaces Native client applications In short: Industrial SCADA Control system framework TANGO Based on CORBA and ZMQ C++ TANGO Java Archiving Python System Centralized config. database TANGO TANGO TANGO TANGO Clients binding binding binding binding (CLI/GUI) Software bus for distributed TANGO software bus objects Provides unified interface to Device Device Device Device Device Device Device Server Server Server Server Server Server Server all equipments hiding how they are HV ps + Pylon OPC UA Data Motion TANGO SNMP connected/managed -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

A Practical Guide for Improving Transparency and Reproducibility in Neuroimaging Research Krzysztof J
bioRxiv preprint first posted online Feb. 12, 2016; doi: http://dx.doi.org/10.1101/039354. The copyright holder for this preprint (which was not peer-reviewed) is the author/funder. It is made available under a CC-BY 4.0 International license. A practical guide for improving transparency and reproducibility in neuroimaging research Krzysztof J. Gorgolewski and Russell A. Poldrack Department of Psychology, Stanford University Abstract Recent years have seen an increase in alarming signals regarding the lack of replicability in neuroscience, psychology, and other related fields. To avoid a widespread crisis in neuroimaging research and consequent loss of credibility in the public eye, we need to improve how we do science. This article aims to be a practical guide for researchers at any stage of their careers that will help them make their research more reproducible and transparent while minimizing the additional effort that this might require. The guide covers three major topics in open science (data, code, and publications) and offers practical advice as well as highlighting advantages of adopting more open research practices that go beyond improved transparency and reproducibility. Introduction The question of how the brain creates the mind has captivated humankind for thousands of years. With recent advances in human in vivo brain imaging, we how have effective tools to peek into biological underpinnings of mind and behavior. Even though we are no longer constrained just to philosophical thought experiments and behavioral observations (which undoubtedly are extremely useful), the question at hand has not gotten any easier. These powerful new tools have largely demonstrated just how complex the biological bases of behavior actually are. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -

Automated Regional Behavioral Analysis for Human Brain Images
METHODS ARTICLE published: 28 August 2012 NEUROINFORMATICS doi: 10.3389/fninf.2012.00023 Automated regional behavioral analysis for human brain images Jack L. Lancaster 1*, Angela R. Laird 1, Simon B. Eickhoff 2,3, Michael J. Martinez 1, P. M i c k l e F o x 1 and Peter T. Fox 1 1 Research Imaging Institute, The University of Texas Health Science Center at San Antonio, San Antonio, TX, USA 2 Institute of Neuroscience and Medicine (INM-1), Research Center, Jülich, Germany 3 Institute for Clinical Neuroscience and Medical Psychology, Heinrich-Heine University, Düsseldorf, Germany Edited by: Behavioral categories of functional imaging experiments along with standardized brain Robert W. Williams, University of coordinates of associated activations were used to develop a method to automate regional Tennessee Health Science Center, behavioral analysis of human brain images. Behavioral and coordinate data were taken USA from the BrainMap database (http://www.brainmap.org/), which documents over 20 years Reviewed by: Glenn D. Rosen, Beth Israel of published functional brain imaging studies. A brain region of interest (ROI) for behavioral Deaconess Medical Center, USA analysis can be defined in functional images, anatomical images or brain atlases, if Khyobeni Mozhui, University of images are spatially normalized to MNI or Talairach standards. Results of behavioral Tennessee Health Science Center, analysis are presented for each of BrainMap’s 51 behavioral sub-domains spanning five USA behavioral domains (Action, Cognition, Emotion, Interoception, and Perception). For each *Correspondence: Jack L. Lancaster, Research Imaging behavioral sub-domain the fraction of coordinates falling within the ROI was computed Institute, The University of Texas and compared with the fraction expected if coordinates for the behavior were not Health Science Center at San clustered, i.e., uniformly distributed. -

Survey of Databases Used in Image Processing and Their Applications
International Journal of Scientific & Engineering Research Volume 2, Issue 10, Oct-2011 1 ISSN 2229-5518 Survey of Databases Used in Image Processing and Their Applications Shubhpreet Kaur, Gagandeep Jindal Abstract- This paper gives review of Medical image database (MIDB) systems which have been developed in the past few years for research for medical fraternity and students. In this paper, I have surveyed all available medical image databases relevant for research and their use. Keywords: Image database, Medical Image Database System. —————————— —————————— 1. INTRODUCTION Measurement and recording techniques, such as electroencephalography, magnetoencephalography Medical imaging is the technique and process used to (MEG), Electrocardiography (EKG) and others, can create images of the human for clinical purposes be seen as forms of medical imaging. Image Analysis (medical procedures seeking to reveal, diagnose or is done to ensure database consistency and reliable examine disease) or medical science. As a discipline, image processing. it is part of biological imaging and incorporates radiology, nuclear medicine, investigative Open source software for medical image analysis radiological sciences, endoscopy, (medical) Several open source software packages are available thermography, medical photography and for performing analysis of medical images: microscopy. ImageJ 3D Slicer ITK Shubhpreet Kaur is currently pursuing masters degree OsiriX program in Computer Science and engineering in GemIdent Chandigarh Engineering College, Mohali, India. E-mail: MicroDicom [email protected] FreeSurfer Gagandeep Jindal is currently assistant processor in 1.1 Images used in Medical Research department Computer Science and Engineering in Here is the description of various modalities that are Chandigarh Engineering College, Mohali, India. E-mail: used for the purpose of research by medical and [email protected] engineering students as well as doctors. -
PDF File, 212 KB
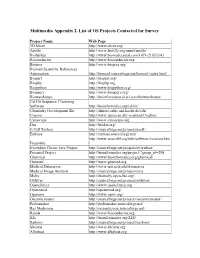
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -

Whole Body Computed Tomography with Advanced Imaging Techniques: a Research Tool for Measuring Body Composition in Dogs
Hindawi Publishing Corporation Journal of Veterinary Medicine Volume 2013, Article ID 610654, 6 pages http://dx.doi.org/10.1155/2013/610654 Research Article Whole Body Computed Tomography with Advanced Imaging Techniques: A Research Tool for Measuring Body Composition in Dogs Dharma Purushothaman,1 Barbara A. Vanselow,2 Shu-Biao Wu,1 Sarah Butler,3 and Wendy Yvonne Brown1 1 School of Environmental and Rural Science, Department of Animal Science, University of New England, Armidale, NSW 2351, Australia 2 NSW Department of Primary Industries, Beef Industry Centre, University of New England, Armidale, NSW 2351, Australia 3 North Hill Vet Clinic, Armidale, NSW 2350, Australia Correspondence should be addressed to Wendy Yvonne Brown; [email protected] Received 6 May 2013; Revised 14 September 2013; Accepted 17 September 2013 Academic Editor: Juan G. Chediack Copyright © 2013 Dharma Purushothaman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The use of computed tomography (CT) to evaluate obesity in canines is limited. Traditional CT image analysis is cumbersome and uses prediction equations that require manual calculations. In order to overcome this, our study investigated the use of advanced image analysis software programs to determine body composition in dogs with an application to canine obesity research. Beagles and greyhounds were chosen for their differences in morphology and propensity to obesity. Whole body CT scans with regular intervals were performed on six beagles and six greyhounds that were subjected to a 28-day weight-gain protocol. -

Mango Ripeness Stages Using Three Image Processing Algorithms
RESEARCH ARTICLE Determining ‘Carabao’ Mango Ripeness Stages Using Three Image Processing Algorithms ٠ Mary Grace C. Galela ٠ Deborah S. Naciongayo ٠ Miguel Carlos S. Guillermo Cinmayii G. Manliguez ٠ Emma Ruth V. Bayogan ٠ Czaryl P. Dioquino University of the Philippines Mindanao, PHILIPPINES Abstract Harvested mangoes are commonly classified or sorted manually. This method is tedious, time consuming, inaccurate and prone to errors. Human inspection is also subjective and factors like visual stress and tiredness may arise that can result in the inconsistencies in judgment. The use of a chroma meter is reliable but the equipment is expensive. This study explored the use of three digital image processing algorithms to determine harvested ‘Carabao’ mango ripeness stages. Canny edge detection, Sobel edge detection, and Laplacian of Gaussian detection algorithms were used to extract a mango image from its original image. The mean red, green, and blue (RGB) values of the detected images were converted to L*a*b* color values which were used to identify the ripeness level of the mango image based on the standard generated from gathered data of ‘Carabao’ mangoes. The standard generated was also based on the mango peel color index scale from University of the Philippines Los Baños Postharvest Horticulture Training and Research Center (PHTRC). The algorithms’ performance had an overall accuracy of 80.5% for canny edge detection algorithm and L*a*b* color extraction using neural networks; 63.88% for Sobel edge detection algorithm and L*a*b* color extraction using rgb2lab function in MATLAB software; and 17.33% Laplacian of Gaussian detection and L*a*b* color extraction using OpenCV. -

Working with the DICOM and Nifti Data Standards in R
JSS Journal of Statistical Software October 2011, Volume 44, Issue 6. http://www.jstatsoft.org/ Working with the DICOM and NIfTI Data Standards in R Brandon Whitcher Volker J. Schmid Andrew Thornton Mango Solutions Ludwig-Maximilians- Cardiff University Universit¨at Munchen¨ Abstract Two packages, oro.dicom and oro.nifti, are provided for the interaction with and manipulation of medical imaging data that conform to the DICOM standard or ANA- LYZE/NIfTI formats. DICOM data, from a single file or directory tree, may be uploaded into R using basic data structures: a data frame for the header information and a matrix for the image data. A list structure is used to organize multiple DICOM files. The S4 class framework is used to develop basic ANALYZE and NIfTI classes, where NIfTI extensions may be used to extend the fixed-byte NIfTI header. One example of this, that has been implemented, is an XML-based \audit trail" tracking the history of operations applied to a data set. The conversion from DICOM to ANALYZE/NIfTI is straightforward using the capabilities of both packages. The S4 classes have been developed to provide a user- friendly interface to the ANALYZE/NIfTI data formats; allowing easy data input, data output, image processing and visualization. Keywords: export, imaging, import, medical, visualization. 1. Introduction Medical imaging is well established in both the clinical and research areas with numerous equipment manufacturers supplying a wide variety of modalities. The DICOM (Digital Imag- ing and Communications in Medicine; http://medical.nema.org/) standard was developed from earlier standards and released in 1993.