Survey of Databases Used in Image Processing and Their Applications
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -
![Downloaded from the Cellprofiler Site [31] to Provide a Starting Point for New Analyses](https://docslib.b-cdn.net/cover/6758/downloaded-from-the-cellprofiler-site-31-to-provide-a-starting-point-for-new-analyses-626758.webp)
Downloaded from the Cellprofiler Site [31] to Provide a Starting Point for New Analyses
Open Access Software2006CarpenteretVolume al. 7, Issue 10, Article R100 CellProfiler: image analysis software for identifying and quantifying comment cell phenotypes Anne E Carpenter*, Thouis R Jones*†, Michael R Lamprecht*, Colin Clarke*†, In Han Kang†, Ola Friman‡, David A Guertin*, Joo Han Chang*, Robert A Lindquist*, Jason Moffat*, Polina Golland† and David M Sabatini*§ reviews Addresses: *Whitehead Institute for Biomedical Research, Cambridge, MA 02142, USA. †Computer Sciences and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02142, USA. ‡Department of Radiology, Brigham and Women's Hospital, Boston, MA 02115, USA. §Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02142, USA. Correspondence: David M Sabatini. Email: [email protected] Published: 31 October 2006 Received: 15 September 2006 Accepted: 31 October 2006 reports Genome Biology 2006, 7:R100 (doi:10.1186/gb-2006-7-10-r100) The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2006/7/10/R100 © 2006 Carpenter et al.; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. deposited research Cell<p>CellProfiler, image analysis the software first free, open-source system for flexible and high-throughput cell image analysis is described.</p> Abstract Biologists can now prepare and image thousands of samples per day using automation, enabling chemical screens and functional genomics (for example, using RNA interference). Here we describe the first free, open-source system designed for flexible, high-throughput cell image analysis, research refereed CellProfiler. -

Sistema De Detección Y Ubicación De Marcadores Visuales Para El Rastreo De Robots Móviles
1 UNIVERSIDAD NACIONAL AUTÓNOMA DE MÉXICO FACULTAD DE INGENIERÍA SISTEMA DE DETECCIÓN Y UBICACIÓN DE MARCADORES VISUALES PARA EL RASTREO DE ROBOTS MÓVILES T E S I S Que para obtener el título de INGENIERO MECATRÓNICO P R E S E N T A EMMANUEL TAPIA BRITO DIRECTOR DE TESIS: DR. VÍCTOR JAVIER GONZÁLEZ VILLELA MÉXICO D.F. JULIO 2013 Dedicatoria A mis padres, por su incondicional apoyo, por ser fuente de inspiración, por su esfuerzo derramado día a día y por aguantarme durante todos estos años. * A Getsemaní por llenar de inmenso amor y alegría mi corazón y por haber compartido conmigo una gran etapa de mi vida. * A Sofía por brindarme tu apoyo y el amor que me dio ánimos para alcanzar esta meta. * A todos mis amigos por haber formado parte de mi vida universitaria. 3 4 Agradecimientos A la Universidad Nacional Autónoma de México por la excelente educación recibida. * A los profesores de la Facultad de Ingeniería, en especial a los del Departamento de Ingeniería Mecatrónica, a quienes debo mi formación profesional. * Al grupo de trabajo MRG, por su soporte técnico y ayuda en la realización de esta Tesis. * Al Dr. Víctor Javier González Villela por la oportunidad, motivación y confianza brindada durante la realización de este proyecto. * A Oscar X. Hurtado Reynoso, por su gran ayuda en la etapa de pruebas con el robot. * A la DGAPA por el apoyo brindado a través del proyecto PAPIIT 1N115811 con título “Investigación y desarrollo en sistemas mecatrónicos: robótica móvil, robótica paralela, robótica híbrida y teleoperación”. 5 6 Resumen Esta tesis presenta los puntos principales del desarrollo de un sistema de visión por computadora con el objetivo de emplearlo como medio de asistencia para el con- trol de robots móviles. -
PDF File, 212 KB
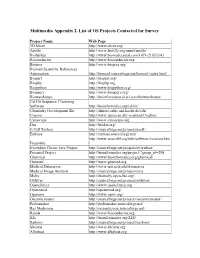
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -

Imagej Basics (Version 1.38)
ImageJ Basics (Version 1.38) ImageJ is a powerful image analysis program that was created at the National Institutes of Health. It is in the public domain, runs on a variety of operating systems and is updated frequently. You may download this program from the source (http://rsb.info.nih.gov/ij/) or copy the ImageJ folder from the C drive of your lab computer. The ImageJ website has instructions for use of the program and links to useful resources. Installing ImageJ on your PC (Windows operating system): Copy the ImageJ folder and transfer it to the C drive of your personal computer. Open the ImageJ folder in the C drive and copy the shortcut (microscope with arrow) to your computer’s desktop. Double click on this desktop shortcut to run ImageJ. See the ImageJ website for Macintosh instructions. ImageJ Window: The ImageJ window will appear on the desktop; do not enlarge this window. Note that this window has a Menu Bar, a Tool Bar and a Status Bar. Menu Bar → Tool Bar → Status Bar → Graphics are from the ImageJ website (http://rsb.info.nih.gov/ij/). Adjusting Memory Allocation: Use the Edit → Options → Memory command to adjust the default memory allocation. Setting the maximum memory value to more than about 75% of real RAM may result in poor performance due to virtual memory "thrashing". Opening an Image File: Select File → Open from the menu bar to open a stored image file. Tool Bar: The various buttons on the tool bar allow you measure, draw, label, fill, etc. A right- click or a double left-click may expand your options with some of the tool buttons. -

Paleoanthropology Society Meeting Abstracts, St. Louis, Mo, 13-14 April 2010
PALEOANTHROPOLOGY SOCIETY MEETING ABSTRACTS, ST. LOUIS, MO, 13-14 APRIL 2010 New Data on the Transition from the Gravettian to the Solutrean in Portuguese Estremadura Francisco Almeida , DIED DEPA, Igespar, IP, PORTUGAL Henrique Matias, Department of Geology, Faculdade de Ciências da Universidade de Lisboa, PORTUGAL Rui Carvalho, Department of Geology, Faculdade de Ciências da Universidade de Lisboa, PORTUGAL Telmo Pereira, FCHS - Departamento de História, Arqueologia e Património, Universidade do Algarve, PORTUGAL Adelaide Pinto, Crivarque. Lda., PORTUGAL From an anthropological perspective, the passage from the Gravettian to the Solutrean is one of the most interesting transition peri- ods in Old World Prehistory. Between 22 kyr BP and 21 kyr BP, during the beginning stages of the Last Glacial Maximum, Iberia and Southwest France witness a process of substitution of a Pan-European Technocomplex—the Gravettian—to one of the first examples of regionalism by Anatomically Modern Humans in the European continent—the Solutrean. While the question of the origins of the Solutrean is almost as old as its first definition, the process under which it substituted the Gravettian started to be readdressed, both in Portugal and in France, after the mid 1990’s. Two chronological models for the transition have been advanced, but until very recently the lack of new archaeological contexts of the period, and the fact that the many of the sequences have been drastically affected by post depositional disturbances during the Lascaux event, prevented their systematic evaluation. Between 2007 and 2009, and in the scope of mitigation projects, archaeological fieldwork has been carried in three open air sites—Terra do Manuel (Rio Maior), Portela 2 (Leiria), and Calvaria 2 (Porto de Mós) whose stratigraphic sequences date precisely to the beginning stages of the LGM. -

Automated Malware Analysis Report for Microdicom-3.0.1
ID: 153765 Sample Name: MicroDicom- 3.0.1-x64.exe Cookbook: default.jbs Time: 13:55:16 Date: 18/07/2019 Version: 26.0.0 Aquamarine Table of Contents Table of Contents 2 Analysis Report MicroDicom-3.0.1-x64.exe 4 Overview 4 General Information 4 Detection 4 Confidence 5 Classification 5 Analysis Advice 5 Mitre Att&ck Matrix 6 Signature Overview 6 AV Detection: 6 Spreading: 6 Networking: 6 Key, Mouse, Clipboard, Microphone and Screen Capturing: 6 System Summary: 7 Data Obfuscation: 7 Persistence and Installation Behavior: 7 Boot Survival: 7 Hooking and other Techniques for Hiding and Protection: 7 Malware Analysis System Evasion: 7 Anti Debugging: 8 HIPS / PFW / Operating System Protection Evasion: 8 Language, Device and Operating System Detection: 8 Behavior Graph 8 Simulations 8 Behavior and APIs 8 Antivirus and Machine Learning Detection 9 Initial Sample 9 Dropped Files 9 Unpacked PE Files 9 Domains 9 URLs 9 Yara Overview 9 Initial Sample 9 PCAP (Network Traffic) 9 Dropped Files 9 Memory Dumps 9 Unpacked PEs 9 Joe Sandbox View / Context 10 IPs 10 Domains 10 ASN 10 JA3 Fingerprints 10 Dropped Files 10 Screenshots 10 Thumbnails 10 Startup 11 Created / dropped Files 11 Domains and IPs 17 Contacted Domains 17 URLs from Memory and Binaries 17 Contacted IPs 17 Static File Info 17 General 18 File Icon 18 Static PE Info 18 General 18 Entrypoint Preview 18 Rich Headers 19 Copyright Joe Security LLC 2019 Page 2 of 44 Data Directories 19 Sections 20 Resources 20 Imports 20 Possible Origin 21 Network Behavior 21 Code Manipulations 21 Statistics 21 -

Anastasia Tyurina [email protected]
1 Anastasia Tyurina [email protected] Summary A specialist in applying or creating mathematical methods to solving problems of developing technologies. A rare expert in solving problems starting from the stage of a stated “word problem” to proof of concept and production software development. Such successful uses of an educational background in mathematics, intellectual courage, and tenacious character include: • developed a unique method of statistical analysis of spectral composition in 1D and 2D stochastic processes for quality control in ultra-precision mirror polishing • developed novel methods of detection, tracking and classification of small moving targets for aerial IR and EO sensors. Used SIFT, and SIRF features, and developed innovative feature-signatures of motion of interest. • developed image processing software for bioinformatics, point source (diffraction objects) detection semiconductor metrology, electron microscopy, failure analysis, diagnostics, system hardware support and pattern recognition • developed statistical software for surface metrology assessment, characterization and generation of statistically similar surfaces to assist development of new optical systems • documented, published and patented original results helping employers technical communications • supported sales with prototypes and presentations • worked well with people – colleagues, customers, researchers, scientists, engineers Tools MATLAB, Octave, OpenCV, ImageJ, Scion Image, Aphelion Image, Gimp, PhotoShop, C/C++, (Visual C environment), GNU development tools, UNIX (Solaris, SGI IRIX), Linux, Windows, MS DOS. Positions and Experience Second Star Algonumerixs – 2008-present, founder and CEO http://www.secondstaralgonumerix.com/ 1) Developed a method of statistical assessment, characterisation and generation of random surface metrology for sper precision X-ray mirror manufacturing in collaboration with Lawrence Berkeley National Laboratory of University of California Berkeley. -

Medical Images Research Framework
Medical Images Research Framework Sabrina Musatian Alexander Lomakin Angelina Chizhova Saint Petersburg State University Saint Petersburg State University Saint Petersburg State University Saint Petersburg, Russia Saint Petersburg, Russia Saint Petersburg, Russia Email: [email protected] Email: [email protected] Email: [email protected] Abstract—with a growing interest in medical research problems for the development of medical instruments and to show and the introduction of machine learning methods for solving successful applications of this library on some real medical those, a need in an environment for integrating modern solu- cases. tions and algorithms into medical applications developed. The main goal of our research is to create medical images research 2. Existing systems for medical image process- framework (MIRF) as a solution for the above problem. MIRF ing is a free open–source platform for the development of medical tools with image processing. We created it to fill in the gap be- There are many open–source packages and software tween innovative research with medical images and integrating systems for working with medical images. Some of them are it into real–world patients treatments workflow. Within a short specifically dedicated for these purposes, others are adapted time, a developer can create a rich medical tool, using MIRF's to be used for medical procedures. modular architecture and a set of included features. MIRF Many of them comprise a set of instruments, dedicated takes the responsibility of handling common functionality for to solving typical tasks, such as images pre–processing medical images processing. The only thing required from the and analysis of the results – ITK [1], visualization – developer is integrating his functionality into a module and VTK [2], real–time pre–processing of images and video – choosing which of the other MIRF's features are needed in the OpenCV [3]. -

Whole Body Computed Tomography with Advanced Imaging Techniques: a Research Tool for Measuring Body Composition in Dogs
Hindawi Publishing Corporation Journal of Veterinary Medicine Volume 2013, Article ID 610654, 6 pages http://dx.doi.org/10.1155/2013/610654 Research Article Whole Body Computed Tomography with Advanced Imaging Techniques: A Research Tool for Measuring Body Composition in Dogs Dharma Purushothaman,1 Barbara A. Vanselow,2 Shu-Biao Wu,1 Sarah Butler,3 and Wendy Yvonne Brown1 1 School of Environmental and Rural Science, Department of Animal Science, University of New England, Armidale, NSW 2351, Australia 2 NSW Department of Primary Industries, Beef Industry Centre, University of New England, Armidale, NSW 2351, Australia 3 North Hill Vet Clinic, Armidale, NSW 2350, Australia Correspondence should be addressed to Wendy Yvonne Brown; [email protected] Received 6 May 2013; Revised 14 September 2013; Accepted 17 September 2013 Academic Editor: Juan G. Chediack Copyright © 2013 Dharma Purushothaman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The use of computed tomography (CT) to evaluate obesity in canines is limited. Traditional CT image analysis is cumbersome and uses prediction equations that require manual calculations. In order to overcome this, our study investigated the use of advanced image analysis software programs to determine body composition in dogs with an application to canine obesity research. Beagles and greyhounds were chosen for their differences in morphology and propensity to obesity. Whole body CT scans with regular intervals were performed on six beagles and six greyhounds that were subjected to a 28-day weight-gain protocol.