Segmentation Tools Frederic Cervenansky What Is Open-Source
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Accurate Segmentation of Brain MR Images
Accurate segmentation of brain MR images Master of Science Thesis in Biomedical Engineering ANTONIO REYES PORRAS PÉREZ Department of Signals and Systems Division of Biomedical Engineering CHALMERS UNIVERSITY OF TECHNOLOGY Göteborg, Sweden, 2010 Report No. EX028/2010 Abstract Full brain segmentation has been of significant interest throughout the years. Recently, many research groups worldwide have been looking into development of patient-specific electromagnetic models for dipole source location in EEG. To obtain this model, accurate segmentation of various tissues and sub-cortical structures is thus required. In this project, the performance of three of the most widely used software packages for brain segmentation has been analyzed: FSL, SPM and FreeSurfer. For the analysis, real images from a patient and a set of phantom images have been used in order to evaluate the performance r of each one of these tools. Keywords: dipole source location, brain, patient-specific model, image segmentation, FSL, SPM, FreeSurfer. Acknowledgements To my advisor, Antony, for his guidance through the project. To my partner, Koushyar, for all the days we have spent in the hospital helping each other. To the staff in Sahlgrenska hospital for their collaboration. To MedTech West for this opportunity to learn. Table of contents 1. Introduction ......................................................................................................................................... 1 2. Magnetic resonance imaging .............................................................................................................. -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -
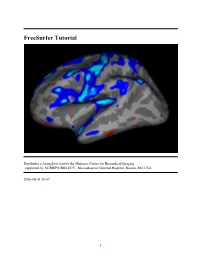
Freesurfer Tutorial
FreeSurfer Tutorial FreeSurfer is brought to you by the Martinos Center for Biomedical Imaging supported by NCRR/P41RR14075, Massachusetts General Hospital, Boston, MA USA 2008-06-01 20:47 1 FreeSurfer Tutorial Table of Contents Section Page Overview and course outline 3 Inspection of Freesurfer output 5 Troubleshooting your output 22 Fixing a bad skull strip 26 Making edits to the white matter 34 Correcting pial surfaces 46 Using control points to fix intensity normalization 50 Talairach registration 55 recon-all: morphometry and reconstruction 69 recon-all: process flow table 97 QDEC Group analysis 100 Group analysis: average subject, design matrix, mri_glmfit 125 Group analysis: visualization and inspection 149 Integrating FreeSurfer and FSL's FEAT 159 Exercise overview 172 Tkmedit reference 176 Tksurfer reference 211 Glossary 227 References 229 Acknowledgments 238 2008-06-01 20:47 2 top FreeSurfer Slides 1. Introduction to Freesurfer - Bruce Fischl 2. Anatomical Analysis with Freesurfer - Doug Greve 3. Surface-based Group Analysis - Doug Greve 4. Applying FreeSurfer Tools to FSL fMRI Analysis - Doug Greve FreeSurfer Tutorial Overview The FreeSurfer tools deal with two main types of data: volumetric data volumes of voxels and surface data polygons that tile a surface. This tutorial should familiarize you with FreeSurfer’s volume and surface processing streams, the recommended workflow to execute these, and many of their component tools. The tutorial also describes some of FreeSurfer's tools for registering volumetric datasets, performing group analysis on morphology data, and integrating FSL Feat output with FreeSurfer overlaying color coded parametric maps onto the cortical surface and visualizing plotted results. -

Confrontsred Tape
IPS: THE MENTAL HEALTH SERVICES CONFERENCE Oct. 8-11, 2015 • New York City When Good Care Confronts Red Tape Navigating the System for Our Patients and Our Practice New York, NY | Sheraton New York Times Square POSTER SESSION OCTOBER 09, 2015 POSTER SESSION 1 P1- 1 ANGIOEDEMA DUE TO POTENTIATING EFFECTS OF RITONIVIR ON RISPERIDONE: CASE REPORT Lead Author: Luisa S. Gonzalez, M.D. Co-Author(s): David A. Kasle MSIII Kavita Kothari M.D. SUMMARY: For many drugs, the liver is the principal site of its metabolism. The most important enzyme system of phase I metabolism is the cytochrome P-450 (CYP450). Ritonavir, a protease inhibitor used for the treatment of human immunodeficiency virus (HIV) infection and acquired immunodeficiency syndrome (AIDS), has been shown to be a potent inhibitor of the (CYP450) 3A and 2D6 isozymes. This inhibition increases the chance for potential drug-drug interactions with compounds that are metabolized by these isoforms. Risperidone, a second generation atypical antipsychotic, is metabolized to a significant extent by the (CYP450) 2D6, and to a lesser extent 3A4. Ritonavir and risperidone can both cause serious, and even life threatening, side effects. An example of one such rare side effect is angioedema which is characterized by edema of the deep dermal and subcutaneous tissues. Here we present three cases of patients residing in a nursing home setting, diagnosed with HIV/AIDS and comorbid psychiatric illness. These patients were all on antiretroviral medications such as ritonavir, and all later presented with varying degrees of edema after long term use of risperidone. We aim to bring to awareness the potential for ritonavir inhibiting the metabolism of risperidone, thus leading to an increased incidence of angioedema in these patients. -

Survey of Databases Used in Image Processing and Their Applications
International Journal of Scientific & Engineering Research Volume 2, Issue 10, Oct-2011 1 ISSN 2229-5518 Survey of Databases Used in Image Processing and Their Applications Shubhpreet Kaur, Gagandeep Jindal Abstract- This paper gives review of Medical image database (MIDB) systems which have been developed in the past few years for research for medical fraternity and students. In this paper, I have surveyed all available medical image databases relevant for research and their use. Keywords: Image database, Medical Image Database System. —————————— —————————— 1. INTRODUCTION Measurement and recording techniques, such as electroencephalography, magnetoencephalography Medical imaging is the technique and process used to (MEG), Electrocardiography (EKG) and others, can create images of the human for clinical purposes be seen as forms of medical imaging. Image Analysis (medical procedures seeking to reveal, diagnose or is done to ensure database consistency and reliable examine disease) or medical science. As a discipline, image processing. it is part of biological imaging and incorporates radiology, nuclear medicine, investigative Open source software for medical image analysis radiological sciences, endoscopy, (medical) Several open source software packages are available thermography, medical photography and for performing analysis of medical images: microscopy. ImageJ 3D Slicer ITK Shubhpreet Kaur is currently pursuing masters degree OsiriX program in Computer Science and engineering in GemIdent Chandigarh Engineering College, Mohali, India. E-mail: MicroDicom [email protected] FreeSurfer Gagandeep Jindal is currently assistant processor in 1.1 Images used in Medical Research department Computer Science and Engineering in Here is the description of various modalities that are Chandigarh Engineering College, Mohali, India. E-mail: used for the purpose of research by medical and [email protected] engineering students as well as doctors. -
PDF File, 212 KB
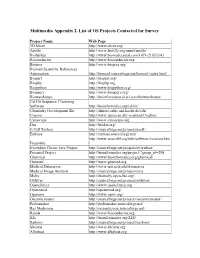
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -

A Novel Mricompatible Brain Ventricle Phantom for Validation of Segmentation and Volumetry Methods
CME JOURNAL OF MAGNETIC RESONANCE IMAGING 36:476–482 (2012) Original Research A Novel MRI-Compatible Brain Ventricle Phantom for Validation of Segmentation and Volumetry Methods Amanda F. Khan, MSc,1,2 John J. Drozd, PhD,2 Robert K. Moreland, MSc,2,3 Robert M. Ta, MSc,1,2 Michael J. Borrie, MB ChB,3,4 Robert Bartha, PhD1,2* and the Alzheimer’s Disease Neuroimaging Initiative Purpose: To create a standardized, MRI-compatible, life- Conclusion: The phantom represents a simple, realistic sized phantom of the brain ventricles to evaluate ventricle and objective method to test the accuracy of lateral ventri- segmentation methods using T1-weighted MRI. An objec- cle segmentation methods and we project it can be tive phantom is needed to test the many different segmen- extended to other anatomical structures. tation programs currently used to measure ventricle vol- Key Words: 3T; brain phantom; MRI; ventricle; software umes in patients with Alzheimer’s disease. validation; segmentation Materials and Methods: A ventricle model was con- J. Magn. Reson. Imaging 2012;36:476–482. structed from polycarbonate using a digital mesh of the VC 2012 Wiley Periodicals, Inc. ventricles created from the 3 Tesla (T) MRI of a subject with Alzheimer’s disease. The ventricle was placed in a brain mold and surrounded with material composed of VOLUMETRY HAS DEMONSTRATED that large mor- 2% agar in water, 0.01% NaCl and 0.0375 mM gadopente- phological changes occur in the brain during the tate dimeglumine to match the signal intensity properties course of Alzheimer’s disease (AD) (1). In particular, of brain tissue in 3T T -weighted MRI. -

Using Camitk for Rapid Prototyping of Interactive Computer Assisted Medical Intervention Applications*
Using CamiTK for Rapid Prototyping of Interactive Computer Assisted Medical Intervention Applications* Emmanuel Promayon, Céline Fouard, Mathieu Bailet, Aurélien Deram, Gaëlle Fiard, Nikolai Hungr, Vincent Luboz, Yohan Payan, Johan Sarrazin, Nicolas Saubat, Sonia Yuki Selmi, Sandrine Voros, Philippe Cinquin and Jocelyne Troccaz Abstract— Computer Assisted Medical Intervention (CAMI hereafter) is a complex multi-disciplinary field. CAMI research requires the collaboration of experts in several fields as diverse as medicine, computer science, mathematics, instrumentation, signal processing, mechanics, modeling, automatics, optics, etc. CamiTK1 is a modular framework that helps researchers and clinicians to collaborate together in order to prototype CAMI applications by regrouping the knowledge and expertise from each discipline. It is an open-source, cross-platform generic and modular tool written in C++ which can handle medical images, surgical navigation, biomedicals simulations and robot control. This paper presents the Computer Assisted Medical Intervention ToolKit (CamiTK) and how it is used in various applications in our research team. Figure 1. Using CamiTK in the CAMI perception – I. INTRODUCTION decision – action loop. The field of CAMI emerged about 30 years ago from the A) Related Work converging evolution of medicine, physics, materials, Numerous programming frameworks exist, generally electronics, computer science and robotics. CAMI aims at focusing on one or two fields of CAMI. For example, some of providing tools that allow the clinician to use multi-modal the most popular free and open-source libraries or data in a rational and quantitative way in order to plan, applications for scientific visualization [2] are Vtk2, simulate and accurately and safely execute mini-invasive Paraview2, VolView2, as well as proprietary software such as medical interventions. -

Medical Images Research Framework
Medical Images Research Framework Sabrina Musatian Alexander Lomakin Angelina Chizhova Saint Petersburg State University Saint Petersburg State University Saint Petersburg State University Saint Petersburg, Russia Saint Petersburg, Russia Saint Petersburg, Russia Email: [email protected] Email: [email protected] Email: [email protected] Abstract—with a growing interest in medical research problems for the development of medical instruments and to show and the introduction of machine learning methods for solving successful applications of this library on some real medical those, a need in an environment for integrating modern solu- cases. tions and algorithms into medical applications developed. The main goal of our research is to create medical images research 2. Existing systems for medical image process- framework (MIRF) as a solution for the above problem. MIRF ing is a free open–source platform for the development of medical tools with image processing. We created it to fill in the gap be- There are many open–source packages and software tween innovative research with medical images and integrating systems for working with medical images. Some of them are it into real–world patients treatments workflow. Within a short specifically dedicated for these purposes, others are adapted time, a developer can create a rich medical tool, using MIRF's to be used for medical procedures. modular architecture and a set of included features. MIRF Many of them comprise a set of instruments, dedicated takes the responsibility of handling common functionality for to solving typical tasks, such as images pre–processing medical images processing. The only thing required from the and analysis of the results – ITK [1], visualization – developer is integrating his functionality into a module and VTK [2], real–time pre–processing of images and video – choosing which of the other MIRF's features are needed in the OpenCV [3]. -

Whole Body Computed Tomography with Advanced Imaging Techniques: a Research Tool for Measuring Body Composition in Dogs
Hindawi Publishing Corporation Journal of Veterinary Medicine Volume 2013, Article ID 610654, 6 pages http://dx.doi.org/10.1155/2013/610654 Research Article Whole Body Computed Tomography with Advanced Imaging Techniques: A Research Tool for Measuring Body Composition in Dogs Dharma Purushothaman,1 Barbara A. Vanselow,2 Shu-Biao Wu,1 Sarah Butler,3 and Wendy Yvonne Brown1 1 School of Environmental and Rural Science, Department of Animal Science, University of New England, Armidale, NSW 2351, Australia 2 NSW Department of Primary Industries, Beef Industry Centre, University of New England, Armidale, NSW 2351, Australia 3 North Hill Vet Clinic, Armidale, NSW 2350, Australia Correspondence should be addressed to Wendy Yvonne Brown; [email protected] Received 6 May 2013; Revised 14 September 2013; Accepted 17 September 2013 Academic Editor: Juan G. Chediack Copyright © 2013 Dharma Purushothaman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The use of computed tomography (CT) to evaluate obesity in canines is limited. Traditional CT image analysis is cumbersome and uses prediction equations that require manual calculations. In order to overcome this, our study investigated the use of advanced image analysis software programs to determine body composition in dogs with an application to canine obesity research. Beagles and greyhounds were chosen for their differences in morphology and propensity to obesity. Whole body CT scans with regular intervals were performed on six beagles and six greyhounds that were subjected to a 28-day weight-gain protocol. -

9. Biomedical Imaging Informatics Daniel L
9. Biomedical Imaging Informatics Daniel L. Rubin, Hayit Greenspan, and James F. Brinkley After reading this chapter, you should know the answers to these questions: 1. What makes images a challenging type of data to be processed by computers, as opposed to non-image clinical data? 2. Why are there many different imaging modalities, and by what major two characteristics do they differ? 3. How are visual and knowledge content in images represented computationally? How are these techniques similar to representation of non-image biomedical data? 4. What sort of applications can be developed to make use of the semantic image content made accessible using the Annotation and Image Markup model? 5. Describe four different types of image processing methods. Why are such methods assembled into a pipeline when creating imaging applications? 6. Give an example of an imaging modality with high spatial resolution. Give an example of a modality that provides functional information. Why are most imaging modalities not capable of providing both? 7. What is the goal in performing segmentation in image analysis? Why is there more than one segmentation method? 8. What are two types of quantitative information in images? What are two types of semantic information in images? How might this information be used in medical applications? 9. What is the difference between image registration and image fusion? Given an example of each. 1 9.1. Introduction Imaging plays a central role in the healthcare process. Imaging is crucial not only to health care, but also to medical communication and education, as well as in research.