3D Slicer Documentation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

Examples of the Electronic Health Record (EHR)
Examples of the Electronic Health Record (EHR) What is Biomedical & Health Informatics? William Hersh Copyright 2020 Oregon Health & Science University Examples of the EHR • Using the Veterans Health Information Systems and Technology Architecture (VistA) • Why VistA? – State-of-the-art EHR that has transformed healthcare in the Veteran’s Health Administration (VHA) (Perlin, 2006; Byrne, 2010) – Not that pretty, but has all of the modern features of the EHR, e.g., clinical decision support (CDS), computerized provider order entry (CPOE), etc. – Distributed under open-source model, unlike most other vendors who do not even allow screen shots to be shown outside their customers’ institutions • Fascinating story (Allen, 2017), but VistA now being phased out in favor of Cerner EHR adopted by Department of Defense (VA, 2017) – https://www.ehrm.va.gov/ WhatIs5 2 1 Some details about VistA • Server written in M (formerly called MUMPS), accessed via command-line interface – Runs in commercial Intersystems Cache (on many platforms) or open-source GT.M (Linux only) • Client (called CPRS) written in Delphi and providers graphical user interface – Only runs on Windows (just about all versions) WhatIs5 3 Logging on to CPRS, the front end to VistA WhatIs5 4 2 Choosing a patient WhatIs5 5 Cover sheet – overview WhatIs5 6 3 Drilling down to details of a problem WhatIs5 7 Details of an allergy WhatIs5 8 4 Viewing vital signs over time WhatIs5 9 More details on problems WhatIs5 10 5 List of active orders WhatIs5 11 Viewing the patient’s notes WhatIs5 12 -

Other Departments and Institutes Courses 1
Other Departments and Institutes Courses 1 Other Departments and Institutes Courses About Course Numbers: 02-250 Introduction to Computational Biology Each Carnegie Mellon course number begins with a two-digit prefix that Spring: 12 units designates the department offering the course (i.e., 76-xxx courses are This class provides a general introduction to computational tools for biology. offered by the Department of English). Although each department maintains The course is divided into two halves. The first half covers computational its own course numbering practices, typically, the first digit after the prefix molecular biology and genomics. It examines important sources of biological indicates the class level: xx-1xx courses are freshmen-level, xx-2xx courses data, how they are archived and made available to researchers, and what are sophomore level, etc. Depending on the department, xx-6xx courses computational tools are available to use them effectively in research. may be either undergraduate senior-level or graduate-level, and xx-7xx In the process, it covers basic concepts in statistics, mathematics, and courses and higher are graduate-level. Consult the Schedule of Classes computer science needed to effectively use these resources and understand (https://enr-apps.as.cmu.edu/open/SOC/SOCServlet/) each semester for their results. Specific topics covered include sequence data, searching course offerings and for any necessary pre-requisites or co-requisites. and alignment, structural data, genome sequencing, genome analysis, genetic variation, gene and protein expression, and biological networks and pathways. The second half covers computational cell biology, including biological modeling and image analysis. It includes homework requiring Computational Biology Courses modification of scripts to perform computational analyses. -

A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours
A 3D interactive multi-object segmentation tool using local robust statistics driven active contours The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Gao, Yi, Ron Kikinis, Sylvain Bouix, Martha Shenton, and Allen Tannenbaum. 2012. A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours. Medical Image Analysis 16, no. 6: 1216–1227. doi:10.1016/j.media.2012.06.002. Published Version doi:10.1016/j.media.2012.06.002 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:28548930 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA NIH Public Access Author Manuscript Med Image Anal. Author manuscript; available in PMC 2013 August 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Med Image Anal. 2012 August ; 16(6): 1216–1227. doi:10.1016/j.media.2012.06.002. A 3D Interactive Multi-object Segmentation Tool using Local Robust Statistics Driven Active Contours Yi Gaoa,*, Ron Kikinisb, Sylvain Bouixa, Martha Shentona, and Allen Tannenbaumc aPsychiatry Neuroimaging Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 bSurgical Planning Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 cDepartments of Electrical and Computer Engineering and Biomedical Engineering, Boston University, Boston, MA 02115 Abstract Extracting anatomical and functional significant structures renders one of the important tasks for both the theoretical study of the medical image analysis, and the clinical and practical community. -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -

Debian Med Integrated Software Environment for All Medical Applications
Debian Med Integrated software environment for all medical applications Andreas Tille 27. February 2013 When people hear for the first time the term ‘Debian Med’ there are usually two kinds of misconceptions. Let us dispel these in advance, so as to clarify subsequent discussion of the project. People familiar with Debian as a large distribution of Free Software usually imag- ine Debian Med to be some kind of customised derivative of Debian tailored for use in a medical environment. Astonishingly, the idea that such customisation can be done entirely within Debian itself is not well known and the technical term Debian Pure Blend seems to be sufficiently unknown outside of the Debian milieu that many people fail to appreciate the concept correctly. There are no separate repositories like Personal Package Archives (PPA) as introduced by Ubuntu for additional software not belong- ing to the official distribution or something like that – a Debian Pure Blend (as the term ’pure’ implies) is Debian itself and if you have received Debian you have full De- bian Med at your disposal. There are other Blends inside Debian like Debian Science, Debian Edu, Debian GIS and others. People working in the health care professions sometimes acquire another miscon- ception about Debian Med, namely that Debian Med is some kind of software primarily dedicated to managing a doctor’s practice. Sometimes people even assume that people assume the Debian Med team actually develops this software. However, the truth about the Debian Med team is that we are a group of Debian developers hard at work incor- porating existing medical software right into the Debian distribution. -

Survey of Databases Used in Image Processing and Their Applications
International Journal of Scientific & Engineering Research Volume 2, Issue 10, Oct-2011 1 ISSN 2229-5518 Survey of Databases Used in Image Processing and Their Applications Shubhpreet Kaur, Gagandeep Jindal Abstract- This paper gives review of Medical image database (MIDB) systems which have been developed in the past few years for research for medical fraternity and students. In this paper, I have surveyed all available medical image databases relevant for research and their use. Keywords: Image database, Medical Image Database System. —————————— —————————— 1. INTRODUCTION Measurement and recording techniques, such as electroencephalography, magnetoencephalography Medical imaging is the technique and process used to (MEG), Electrocardiography (EKG) and others, can create images of the human for clinical purposes be seen as forms of medical imaging. Image Analysis (medical procedures seeking to reveal, diagnose or is done to ensure database consistency and reliable examine disease) or medical science. As a discipline, image processing. it is part of biological imaging and incorporates radiology, nuclear medicine, investigative Open source software for medical image analysis radiological sciences, endoscopy, (medical) Several open source software packages are available thermography, medical photography and for performing analysis of medical images: microscopy. ImageJ 3D Slicer ITK Shubhpreet Kaur is currently pursuing masters degree OsiriX program in Computer Science and engineering in GemIdent Chandigarh Engineering College, Mohali, India. E-mail: MicroDicom [email protected] FreeSurfer Gagandeep Jindal is currently assistant processor in 1.1 Images used in Medical Research department Computer Science and Engineering in Here is the description of various modalities that are Chandigarh Engineering College, Mohali, India. E-mail: used for the purpose of research by medical and [email protected] engineering students as well as doctors. -

An Open-Source Research Platform for Image-Guided Therapy
Int J CARS DOI 10.1007/s11548-015-1292-0 ORIGINAL ARTICLE CustusX: an open-source research platform for image-guided therapy Christian Askeland1,3 · Ole Vegard Solberg1 · Janne Beate Lervik Bakeng1 · Ingerid Reinertsen1 · Geir Arne Tangen1 · Erlend Fagertun Hofstad1 · Daniel Høyer Iversen1,2,3 · Cecilie Våpenstad1,2 · Tormod Selbekk1,3 · Thomas Langø1,3 · Toril A. Nagelhus Hernes2,3 · Håkon Olav Leira2,3 · Geirmund Unsgård2,3 · Frank Lindseth1,2,3 Received: 3 July 2015 / Accepted: 31 August 2015 © The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Results The validation experiments show a navigation sys- Purpose CustusX is an image-guided therapy (IGT) research tem accuracy of <1.1mm, a frame rate of 20 fps, and latency platform dedicated to intraoperative navigation and ultra- of 285ms for a typical setup. The current platform is exten- sound imaging. In this paper, we present CustusX as a robust, sible, user-friendly and has a streamlined architecture and accurate, and extensible platform with full access to data and quality process. CustusX has successfully been used for algorithms and show examples of application in technologi- IGT research in neurosurgery, laparoscopic surgery, vascular cal and clinical IGT research. surgery, and bronchoscopy. Methods CustusX has been developed continuously for Conclusions CustusX is now a mature research platform more than 15years based on requirements from clinical for intraoperative navigation and ultrasound imaging and is and technological researchers within the framework of a ready for use by the IGT research community. CustusX is well-defined software quality process. The platform was open-source and freely available at http://www.custusx.org. -

Medical Image Processing Software
Wohlers Report 2018 Medical Image Processing Software Medical image Patient-specific medical devices and anatomical models are almost always produced using radiological imaging data. Medical image processing processing software is used to translate between radiology file formats and various software AM file formats. Theoretically, any volumetric radiological imaging dataset by Andy Christensen could be used to create these devices and models. However, without high- and Nicole Wake quality medical image data, the output from AM can be less than ideal. In this field, the old adage of “garbage in, garbage out” definitely applies. Due to the relative ease of image post-processing, computed tomography (CT) is the usual method for imaging bone structures and contrast- enhanced vasculature. In the dental field and for oral- and maxillofacial surgery, in-office cone-beam computed tomography (CBCT) has become popular. Another popular imaging technique that can be used to create anatomical models is magnetic resonance imaging (MRI). MRI is less useful for bone imaging, but its excellent soft tissue contrast makes it useful for soft tissue structures, solid organs, and cancerous lesions. Computed tomography: CT uses many X-ray projections through a subject to computationally reconstruct a cross-sectional image. As with traditional 2D X-ray imaging, a narrow X-ray beam is directed to pass through the subject and project onto an opposing detector. To create a cross-sectional image, the X-ray source and detector rotate around a stationary subject and acquire images at a number of angles. An image of the cross-section is then computed from these projections in a post-processing step. -
PDF File, 212 KB
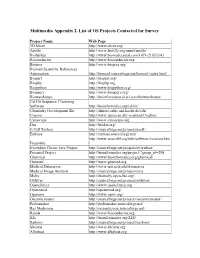
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -

Comparison of Open-Source Electronic Health Record Systems Based on Functional and User Performance Criteria
Original Article Healthc Inform Res. 2019 April;25(2):89-98. https://doi.org/10.4258/hir.2019.25.2.89 pISSN 2093-3681 • eISSN 2093-369X Comparison of Open-Source Electronic Health Record Systems Based on Functional and User Performance Criteria Saptarshi Purkayastha1, Roshini Allam1, Pallavi Maity1, Judy W. Gichoya2 1Department of BioHealth Informatics, Indiana University–Purdue University Indianapolis, Indianapolis, IN, USA 2Dotter Institute, Oregon Health and Science University, Portland, OR, USA Objectives: Open-source Electronic Health Record (EHR) systems have gained importance. The main aim of our research is to guide organizational choice by comparing the features, functionality, and user-facing system performance of the five most popular open-source EHR systems. Methods: We performed qualitative content analysis with a directed approach on recently published literature (2012–2017) to develop an integrated set of criteria to compare the EHR systems. The functional criteria are an integration of the literature, meaningful use criteria, and the Institute of Medicine’s functional requirements of EHR, whereas the user-facing system performance is based on the time required to perform basic tasks within the EHR system. Results: Based on the Alexa web ranking and Google Trends, the five most popular EHR systems at the time of our study were OSHERA VistA, GNU Health, the Open Medical Record System (OpenMRS), Open Electronic Medical Record (OpenEMR), and OpenEHR. We also found the trends in popularity of the EHR systems and the locations where they were more popular than others. OpenEMR met all the 32 functional criteria, OSHERA VistA met 28, OpenMRS met 12 fully and 11 partially, OpenEHR-based EHR met 10 fully and 3 partially, and GNU Health met the least with only 10 criteria fully and 2 partially.