Designing Customized 3D Printed Models for Surgical Planning in Repair of Congenital Heart Defects
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Management of Large Sets of Image Data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak
Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Karol Kozak Download free books at Karol Kozak Management of large sets of image data Capture, Databases, Image Processing, Storage, Visualization Download free eBooks at bookboon.com 2 Management of large sets of image data: Capture, Databases, Image Processing, Storage, Visualization 1st edition © 2014 Karol Kozak & bookboon.com ISBN 978-87-403-0726-9 Download free eBooks at bookboon.com 3 Management of large sets of image data Contents Contents 1 Digital image 6 2 History of digital imaging 10 3 Amount of produced images – is it danger? 18 4 Digital image and privacy 20 5 Digital cameras 27 5.1 Methods of image capture 31 6 Image formats 33 7 Image Metadata – data about data 39 8 Interactive visualization (IV) 44 9 Basic of image processing 49 Download free eBooks at bookboon.com 4 Click on the ad to read more Management of large sets of image data Contents 10 Image Processing software 62 11 Image management and image databases 79 12 Operating system (os) and images 97 13 Graphics processing unit (GPU) 100 14 Storage and archive 101 15 Images in different disciplines 109 15.1 Microscopy 109 360° 15.2 Medical imaging 114 15.3 Astronomical images 117 15.4 Industrial imaging 360° 118 thinking. 16 Selection of best digital images 120 References: thinking. 124 360° thinking . 360° thinking. Discover the truth at www.deloitte.ca/careers Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. Discover the truth at www.deloitte.ca/careers © Deloitte & Touche LLP and affiliated entities. -

A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours
A 3D interactive multi-object segmentation tool using local robust statistics driven active contours The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Gao, Yi, Ron Kikinis, Sylvain Bouix, Martha Shenton, and Allen Tannenbaum. 2012. A 3D Interactive Multi-Object Segmentation Tool Using Local Robust Statistics Driven Active Contours. Medical Image Analysis 16, no. 6: 1216–1227. doi:10.1016/j.media.2012.06.002. Published Version doi:10.1016/j.media.2012.06.002 Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:28548930 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA NIH Public Access Author Manuscript Med Image Anal. Author manuscript; available in PMC 2013 August 01. NIH-PA Author ManuscriptPublished NIH-PA Author Manuscript in final edited NIH-PA Author Manuscript form as: Med Image Anal. 2012 August ; 16(6): 1216–1227. doi:10.1016/j.media.2012.06.002. A 3D Interactive Multi-object Segmentation Tool using Local Robust Statistics Driven Active Contours Yi Gaoa,*, Ron Kikinisb, Sylvain Bouixa, Martha Shentona, and Allen Tannenbaumc aPsychiatry Neuroimaging Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 bSurgical Planning Laboratory, Brigham & Women's Hospital, Harvard Medical School, Boston, MA 02115 cDepartments of Electrical and Computer Engineering and Biomedical Engineering, Boston University, Boston, MA 02115 Abstract Extracting anatomical and functional significant structures renders one of the important tasks for both the theoretical study of the medical image analysis, and the clinical and practical community. -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -

Survey of Databases Used in Image Processing and Their Applications
International Journal of Scientific & Engineering Research Volume 2, Issue 10, Oct-2011 1 ISSN 2229-5518 Survey of Databases Used in Image Processing and Their Applications Shubhpreet Kaur, Gagandeep Jindal Abstract- This paper gives review of Medical image database (MIDB) systems which have been developed in the past few years for research for medical fraternity and students. In this paper, I have surveyed all available medical image databases relevant for research and their use. Keywords: Image database, Medical Image Database System. —————————— —————————— 1. INTRODUCTION Measurement and recording techniques, such as electroencephalography, magnetoencephalography Medical imaging is the technique and process used to (MEG), Electrocardiography (EKG) and others, can create images of the human for clinical purposes be seen as forms of medical imaging. Image Analysis (medical procedures seeking to reveal, diagnose or is done to ensure database consistency and reliable examine disease) or medical science. As a discipline, image processing. it is part of biological imaging and incorporates radiology, nuclear medicine, investigative Open source software for medical image analysis radiological sciences, endoscopy, (medical) Several open source software packages are available thermography, medical photography and for performing analysis of medical images: microscopy. ImageJ 3D Slicer ITK Shubhpreet Kaur is currently pursuing masters degree OsiriX program in Computer Science and engineering in GemIdent Chandigarh Engineering College, Mohali, India. E-mail: MicroDicom [email protected] FreeSurfer Gagandeep Jindal is currently assistant processor in 1.1 Images used in Medical Research department Computer Science and Engineering in Here is the description of various modalities that are Chandigarh Engineering College, Mohali, India. E-mail: used for the purpose of research by medical and [email protected] engineering students as well as doctors. -

An Open-Source Research Platform for Image-Guided Therapy
Int J CARS DOI 10.1007/s11548-015-1292-0 ORIGINAL ARTICLE CustusX: an open-source research platform for image-guided therapy Christian Askeland1,3 · Ole Vegard Solberg1 · Janne Beate Lervik Bakeng1 · Ingerid Reinertsen1 · Geir Arne Tangen1 · Erlend Fagertun Hofstad1 · Daniel Høyer Iversen1,2,3 · Cecilie Våpenstad1,2 · Tormod Selbekk1,3 · Thomas Langø1,3 · Toril A. Nagelhus Hernes2,3 · Håkon Olav Leira2,3 · Geirmund Unsgård2,3 · Frank Lindseth1,2,3 Received: 3 July 2015 / Accepted: 31 August 2015 © The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Results The validation experiments show a navigation sys- Purpose CustusX is an image-guided therapy (IGT) research tem accuracy of <1.1mm, a frame rate of 20 fps, and latency platform dedicated to intraoperative navigation and ultra- of 285ms for a typical setup. The current platform is exten- sound imaging. In this paper, we present CustusX as a robust, sible, user-friendly and has a streamlined architecture and accurate, and extensible platform with full access to data and quality process. CustusX has successfully been used for algorithms and show examples of application in technologi- IGT research in neurosurgery, laparoscopic surgery, vascular cal and clinical IGT research. surgery, and bronchoscopy. Methods CustusX has been developed continuously for Conclusions CustusX is now a mature research platform more than 15years based on requirements from clinical for intraoperative navigation and ultrasound imaging and is and technological researchers within the framework of a ready for use by the IGT research community. CustusX is well-defined software quality process. The platform was open-source and freely available at http://www.custusx.org. -

Medical Image Processing Software
Wohlers Report 2018 Medical Image Processing Software Medical image Patient-specific medical devices and anatomical models are almost always produced using radiological imaging data. Medical image processing processing software is used to translate between radiology file formats and various software AM file formats. Theoretically, any volumetric radiological imaging dataset by Andy Christensen could be used to create these devices and models. However, without high- and Nicole Wake quality medical image data, the output from AM can be less than ideal. In this field, the old adage of “garbage in, garbage out” definitely applies. Due to the relative ease of image post-processing, computed tomography (CT) is the usual method for imaging bone structures and contrast- enhanced vasculature. In the dental field and for oral- and maxillofacial surgery, in-office cone-beam computed tomography (CBCT) has become popular. Another popular imaging technique that can be used to create anatomical models is magnetic resonance imaging (MRI). MRI is less useful for bone imaging, but its excellent soft tissue contrast makes it useful for soft tissue structures, solid organs, and cancerous lesions. Computed tomography: CT uses many X-ray projections through a subject to computationally reconstruct a cross-sectional image. As with traditional 2D X-ray imaging, a narrow X-ray beam is directed to pass through the subject and project onto an opposing detector. To create a cross-sectional image, the X-ray source and detector rotate around a stationary subject and acquire images at a number of angles. An image of the cross-section is then computed from these projections in a post-processing step. -
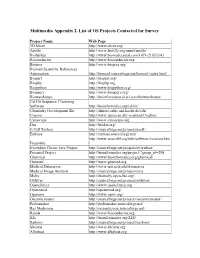
PDF File, 212 KB
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/ -

Whole Body Computed Tomography with Advanced Imaging Techniques: a Research Tool for Measuring Body Composition in Dogs
Hindawi Publishing Corporation Journal of Veterinary Medicine Volume 2013, Article ID 610654, 6 pages http://dx.doi.org/10.1155/2013/610654 Research Article Whole Body Computed Tomography with Advanced Imaging Techniques: A Research Tool for Measuring Body Composition in Dogs Dharma Purushothaman,1 Barbara A. Vanselow,2 Shu-Biao Wu,1 Sarah Butler,3 and Wendy Yvonne Brown1 1 School of Environmental and Rural Science, Department of Animal Science, University of New England, Armidale, NSW 2351, Australia 2 NSW Department of Primary Industries, Beef Industry Centre, University of New England, Armidale, NSW 2351, Australia 3 North Hill Vet Clinic, Armidale, NSW 2350, Australia Correspondence should be addressed to Wendy Yvonne Brown; [email protected] Received 6 May 2013; Revised 14 September 2013; Accepted 17 September 2013 Academic Editor: Juan G. Chediack Copyright © 2013 Dharma Purushothaman et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. The use of computed tomography (CT) to evaluate obesity in canines is limited. Traditional CT image analysis is cumbersome and uses prediction equations that require manual calculations. In order to overcome this, our study investigated the use of advanced image analysis software programs to determine body composition in dogs with an application to canine obesity research. Beagles and greyhounds were chosen for their differences in morphology and propensity to obesity. Whole body CT scans with regular intervals were performed on six beagles and six greyhounds that were subjected to a 28-day weight-gain protocol. -

An Open Source Freeware Software for Ultrasound Imaging and Elastography
Proceedings of the eNTERFACE’07 Workshop on Multimodal Interfaces, Istanbul,˙ Turkey, July 16 - August 10, 2007 USIMAGTOOL: AN OPEN SOURCE FREEWARE SOFTWARE FOR ULTRASOUND IMAGING AND ELASTOGRAPHY Ruben´ Cardenes-Almeida´ 1, Antonio Tristan-Vega´ 1, Gonzalo Vegas-Sanchez-Ferrero´ 1, Santiago Aja-Fernandez´ 1, Veronica´ Garc´ıa-Perez´ 1, Emma Munoz-Moreno˜ 1, Rodrigo de Luis-Garc´ıa 1, Javier Gonzalez-Fern´ andez´ 2, Dar´ıo Sosa-Cabrera 2, Karl Krissian 2, Suzanne Kieffer 3 1 LPI, University of Valladolid, Spain 2 CTM, University of Las Palmas de Gran Canaria 3 TELE Laboratory, Universite´ catholique de Louvain, Louvain-la-Neuve, Belgium ABSTRACT • Open source code: to be able for everyone to modify and reuse the source code. UsimagTool will prepare specific software for the physician to change parameters for filtering and visualization in Ultrasound • Efficiency, robust and fast: using a standard object ori- Medical Imaging in general and in Elastography in particular, ented language such as C++. being the first software tool for researchers and physicians to • Modularity and flexibility for developers: in order to chan- compute elastography with integrated algorithms and modular ge or add functionalities as fast as possible. coding capabilities. It will be ready to implement in different • Multi-platform: able to run in many Operating systems ecographic systems. UsimagTool is based on C++, and VTK/ITK to be useful for more people. functions through a hidden layer, which means that participants may import their own functions and/or use the VTK/ITK func- • Usability: provided with an easy to use GUI to interact tions. as easy as possible with the end user. -

Evaluation of DICOM Viewer Software for Workflow Integration in Clinical Trials
Evaluation of DICOM Viewer Software for Workflow Integration in Clinical Trials Daniel Haak1*, Charles-E. Page, Klaus Kabino, Thomas M. Deserno Department of Medical Informatics, Uniklinik RWTH Aachen, 52057 Aachen, Germany ABSTRACT The digital imaging and communications in medicine (DICOM) protocol is nowadays the leading standard for capture, exchange and storage of image data in medical applications. A broad range of commercial, free, and open source software tools supporting a variety of DICOM functionality exists. However, different from patient’s care in hospital, DICOM has not yet arrived in electronic data capture systems (EDCS) for clinical trials. Due to missing integration, even just the visualization of patient’s image data in electronic case report forms (eCRFs) is impossible. Four increasing levels for integration of DICOM components into EDCS are conceivable, raising functionality but also demands on interfaces with each level. Hence, in this paper, a comprehensive evaluation of 27 DICOM viewer software projects is performed, investigating viewing functionality as well as interfaces for integration. Concerning general, integration, and viewing requirements the survey involves the criteria (i) license, (ii) support, (iii) platform, (iv) interfaces, (v) two- dimensional (2D) and (vi) three-dimensional (3D) image viewing functionality. Optimal viewers are suggested for applications in clinical trials for 3D imaging, hospital communication, and workflow. Focusing on open source solutions, the viewers ImageJ and MicroView are superior for 3D visualization, whereas GingkoCADx is advantageous for hospital integration. Concerning workflow optimization in multi-centered clinical trials, we suggest the open source viewer Weasis. Covering most use cases, an EDCS and PACS interconnection with Weasis is suggested. -
Development and Characterization of the Invesalius Navigator Software for Navigated Transcranial Magnetic Stimulation T ⁎ Victor Hugo Souzaa, ,1, Renan H
Journal of Neuroscience Methods 309 (2018) 109–120 Contents lists available at ScienceDirect Journal of Neuroscience Methods journal homepage: www.elsevier.com/locate/jneumeth Development and characterization of the InVesalius Navigator software for navigated transcranial magnetic stimulation T ⁎ Victor Hugo Souzaa, ,1, Renan H. Matsudaa, André S.C. Peresa,b, Paulo Henrique J. Amorimc, Thiago F. Moraesc, Jorge Vicente L. Silvac, Oswaldo Baffaa a Departamento de Física, Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto, Universidade de São Paulo, Av. Bandeirantes, 3900, 14040-901, Ribeirão Preto, SP, Brazil b Instituto Internacional de Neurociência de Natal Edmond e Lily Safra, Instituto Santos Dumont, Rodovia RN 160 Km 03, 3003, 59280-000, Macaíba, RN, Brazil c Núcleo de Tecnologias Tridimensionais, Centro de Tecnologia da Informação Renato Archer, Rodovia Dom Pedro I Km 143, 13069-901, Campinas, SP, Brazil GRAPHICAL ABSTRACT ARTICLE INFO ABSTRACT Keywords: Background: Neuronavigation provides visual guidance of an instrument during procedures of neurological in- Neuronavigation terventions, and has been shown to be a valuable tool for accurately positioning transcranial magnetic stimu- Transcranial magnetic stimulation lation (TMS) coils relative to an individual’s anatomy. Despite the importance of neuronavigation, its high cost, Localization error low portability, and low availability of magnetic resonance imaging facilities limit its insertion in research and Co-registration clinical environments. Coil positioning New method: We have developed and validated the InVesalius Navigator as the first free, open-source software Surgical planning for image-guided navigated TMS, compatible with multiple tracking devices. A point-based, co-registration al- gorithm and a guiding interface were designed for tracking any instrument (e.g.