Chapter 17: Image Post Processing and Analysis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FACIAL RECOGNITION: a Or FACIALRT RECOGNITION: a Or RT Facial Recognition – Art Or Science?
FACIAL RECOGNITION: A or FACIALRT RECOGNITION: A or RT Facial Recognition – Art or Science? Both the public and law enforcement often misunderstand facial recognition technology. Hollywood would have you believe that with facial recognition technology, law enforcement can identify an individual from an image, while also accessing all of their personal information. The “Big Brother” narrative makes good television, but it vastly misrepresents how law enforcement agencies actually use facial recognition. Additionally, crime dramas like “CSI,” and its endless spinoffs, spin the narrative that law enforcement easily uses facial recognition Roger Rodriguez joined to generate matches. In reality, the facial recognition process Vigilant Solutions after requires a great deal of manual, human analysis and an image of serving over 20 years a certain quality to make a possible match. To date, the quality with the NYPD, where he threshold for images has been hard to reach and has often spearheaded the NYPD’s first frustrated law enforcement looking to generate effective leads. dedicated facial recognition unit. The unit has conducted Think of facial recognition as the 21st-century evolution of the more than 8,500 facial sketch artist. It holds the promise to be much more accurate, recognition investigations, but in the end it is still just the source of a lead that needs to be with over 3,000 possible matches and approximately verified and followed up by intelligent police work. 2,000 arrests. Roger’s enhancement techniques are Today, innovative facial now recognized worldwide recognition technology and have changed law techniques make it possible to enforcement’s approach generate investigative leads to the utilization of facial regardless of image quality. -

Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs
Qualitative Comparison of Conventional and Oblique MRI for Detection of Herniated Spinal Discs Doug Dean Final Project Presentation ENGN 2500: Medical Image Analysis May 16, 2011 Tuesday, May 17, 2011 Outline • Review of the problem presented in the paper: “A comparison of angled sagittal MRI and conventional MRI in the diagnosis of herniated disc and stenosis in the cervical foramen” (Authors: Shim JH, Park CK, Lee JH, et. al) • Approach to solve this problem • Data Acquisition • Analysis Methods • Results • Discussion/Conclusions Tuesday, May 17, 2011 Review of Problem • Difficult to identify herniated discs and spinal stenosis using conventional (2D) MRI techniques • These conventional methods result in patients condition being misdiagnosed. “Conventional MRI”: Images acquired along one of three anatomical planes Tuesday, May 17, 2011 3D reconstructive CT Axial, T2-weighted Image: Image shows that the Cervical Foramen is cervical foramina are directed at 45 degrees directed downward around with respect to coronal 10-15 degrees with plane. respect to axial plane Tuesday, May 17, 2011 Orientation of Images Conventional MRI: Sagittal Protocol Oblique MRI: Sagittal Protocol Tuesday, May 17, 2011 Timeline • Week 1 (4/11-4/16) • Work on developing MR imaging protocols and sequences • Recruit volunteers (~4-5 volunteers) • Week 2 (4/17-4/23) • Continue developing imaging sequences and begin data acquisition at the MRI facility • Assisted by Dr. Deoni • Week 3&4 (4/24-5/7) • Continue developing and testing sequence • 4/27/2011: Acquisition of first subject • Mid Project Presentation: Describe the imaging protocols, present data that had been acquired from previous week, describe what still needs to be done. -

MITK-Modelfit: a Generic Open-Source Framework for Model Fits and Their Exploration in Medical Imaging – Design, Implementatio
MITK-ModelFit: A generic open-source framework for model fits and their exploration in medical imaging – design, implementation and application on the example of DCE-MRI Charlotte Debus1-5,*,#, Ralf Floca5,6,*,#, Michael Ingrisch7, Ina Kompan5,6, Klaus Maier-Hein5,6,8, Amir Abdollahi1-5, and Marco Nolden6 1German Cancer Consortium (DKTK), Heidelberg, Germany 2Translational Radiation Oncology, German Cancer Research Center (DKFZ), Heidelberg, Germany 3Department of Radiation Oncology, Heidelberg Ion-Beam Therapy Center (HIT), Heidelberg University Hospital, Heidelberg, Germany 4National Center for Tumor Diseases (NCT), Heidelberg, Germany 5Heidelberg Institute of Radiation Oncology (HIRO), Germany 6Division of Medical Image Computing, German Cancer Research Center DKFZ, Germany 7Department of Radiology, University Hospital Munich, Ludwig-Maximilians-University Munich, Germany 8Section Pattern Recognition, Department of Radiation Oncology, Heidelberg University Hospital, Heidelberg, Germany # Correspondence: Charlotte Debus, PhD Department of Translational Radiation Oncology Heidelberg Ion-Beam Therapy Center (HIT) Im Neuenheimer Feld 450 69120 Heidelberg, Germany Email: [email protected] Phone: +49 6221 6538281 Ralf Floca, PhD Division of Medical Image Computing German Cancer Research Center (DKFZ) Im Neuenheimer Feld 280 69120 Heidelberg, Germany Email: [email protected] Phone: + 49 6221 42 2560 * Shared first-authors 1 Abstract Background: Many medical imaging techniques utilize fitting approaches for quantitative parameter estimation and analysis. Common examples are pharmacokinetic modeling in dynamic contrast- enhanced (DCE) magnetic resonance imaging (MRI)/computed tomography (CT), apparent diffusion coefficient calculations and intravoxel incoherent motion modeling in diffusion-weighted MRI and Z- spectra analysis in chemical exchange saturation transfer MRI. Most available software tools are limited to a special purpose and do not allow for own developments and extensions. -

An Introduction to Image Analysis Using Imagej
An introduction to image analysis using ImageJ Mark Willett, Imaging and Microscopy Centre, Biological Sciences, University of Southampton. Pete Johnson, Biophotonics lab, Institute for Life Sciences University of Southampton. 1 “Raw Images, regardless of their aesthetics, are generally qualitative and therefore may have limited scientific use”. “We may need to apply quantitative methods to extrapolate meaningful information from images”. 2 Examples of statistics that can be extracted from image sets . Intensities (FRET, channel intensity ratios, target expression levels, phosphorylation etc). Object counts e.g. Number of cells or intracellular foci in an image. Branch counts and orientations in branching structures. Polarisations and directionality . Colocalisation of markers between channels that may be suggestive of structure or multiple target interactions. Object Clustering . Object Tracking in live imaging data. 3 Regardless of the image analysis software package or code that you use….. • ImageJ, Fiji, Matlab, Volocity and IMARIS apps. • Java and Python coding languages. ….image analysis comprises of a workflow of predefined functions which can be native, user programmed, downloaded as plugins or even used between apps. This is much like a flow diagram or computer code. 4 Here’s one example of an image analysis workflow: Apply ROI Choose Make Acquisition Processing to original measurement measurements image type(s) Thresholding Save to ROI manager Make binary mask Make ROI from binary using “Create selection” Calculate x̄, Repeat n Chart data SD, TTEST times and interpret and Δ 5 A few example Functions that can inserted into an image analysis workflow. You can mix and match them to achieve the analysis that you want. -

A Review on Image Segmentation Clustering Algorithms Devarshi Naik , Pinal Shah
Devarshi Naik et al, / (IJCSIT) International Journal of Computer Science and Information Technologies, Vol. 5 (3) , 2014, 3289 - 3293 A Review on Image Segmentation Clustering Algorithms Devarshi Naik , Pinal Shah Department of Information Technology, Charusat University CSPIT, Changa, di.Anand, GJ,India Abstract— Clustering attempts to discover the set of consequential groups where those within each group are more closely related to one another than the others assigned to different groups. Image segmentation is one of the most important precursors for image processing–based applications and has a crucial impact on the overall performance of the developed systems. Robust segmentation has been the subject of research for many years, but till now published work indicates that most of the developed image segmentation algorithms have been designed in conjunction with particular applications. The aim of the segmentation process consists of dividing the input image into several disjoint regions with similar characteristics such as colour and texture. Keywords— Clustering, K-means, Fuzzy C-means, Expectation Maximization, Self organizing map, Hierarchical, Graph Theoretic approach. Fig. 1 Similar data points grouped together into clusters I. INTRODUCTION II. SEGMENTATION ALGORITHMS Images are considered as one of the most important Image segmentation is the first step in image analysis medium of conveying information. The main idea of the and pattern recognition. It is a critical and essential image segmentation is to group pixels in homogeneous component of image analysis system, is one of the most regions and the usual approach to do this is by ‘common difficult tasks in image processing, and determines the feature. Features can be represented by the space of colour, quality of the final result of analysis. -

Image Analysis for Face Recognition
Image Analysis for Face Recognition Xiaoguang Lu Dept. of Computer Science & Engineering Michigan State University, East Lansing, MI, 48824 Email: [email protected] Abstract In recent years face recognition has received substantial attention from both research com- munities and the market, but still remained very challenging in real applications. A lot of face recognition algorithms, along with their modifications, have been developed during the past decades. A number of typical algorithms are presented, being categorized into appearance- based and model-based schemes. For appearance-based methods, three linear subspace analysis schemes are presented, and several non-linear manifold analysis approaches for face recognition are briefly described. The model-based approaches are introduced, including Elastic Bunch Graph matching, Active Appearance Model and 3D Morphable Model methods. A number of face databases available in the public domain and several published performance evaluation results are digested. Future research directions based on the current recognition results are pointed out. 1 Introduction In recent years face recognition has received substantial attention from researchers in biometrics, pattern recognition, and computer vision communities [1][2][3][4]. The machine learning and com- puter graphics communities are also increasingly involved in face recognition. This common interest among researchers working in diverse fields is motivated by our remarkable ability to recognize peo- ple and the fact that human activity is a primary concern both in everyday life and in cyberspace. Besides, there are a large number of commercial, security, and forensic applications requiring the use of face recognition technologies. These applications include automated crowd surveillance, ac- cess control, mugshot identification (e.g., for issuing driver licenses), face reconstruction, design of human computer interface (HCI), multimedia communication (e.g., generation of synthetic faces), 1 and content-based image database management. -

Analysis of Artificial Intelligence Based Image Classification Techniques Dr
Journal of Innovative Image Processing (JIIP) (2020) Vol.02/ No. 01 Pages: 44-54 https://www.irojournals.com/iroiip/ DOI: https://doi.org/10.36548/jiip.2020.1.005 Analysis of Artificial Intelligence based Image Classification Techniques Dr. Subarna Shakya Professor, Department of Electronics and Computer Engineering, Central Campus, Institute of Engineering, Pulchowk, Tribhuvan University. Email: [email protected]. Abstract: Time is an essential resource for everyone wants to save in their life. The development of technology inventions made this possible up to certain limit. Due to life style changes people are purchasing most of their needs on a single shop called super market. As the purchasing item numbers are huge, it consumes lot of time for billing process. The existing billing systems made with bar code reading were able to read the details of certain manufacturing items only. The vegetables and fruits are not coming with a bar code most of the time. Sometimes the seller has to weight the items for fixing barcode before the billing process or the biller has to type the item name manually for billing process. This makes the work double and consumes lot of time. The proposed artificial intelligence based image classification system identifies the vegetables and fruits by seeing through a camera for fast billing process. The proposed system is validated with its accuracy over the existing classifiers Support Vector Machine (SVM), K-Nearest Neighbor (KNN), Random Forest (RF) and Discriminant Analysis (DA). Keywords: Fruit image classification, Automatic billing system, Image classifiers, Computer vision recognition. 1. Introduction Artificial intelligences are given to a machine to work on its task independently without need of any manual guidance. -

Survey of Databases Used in Image Processing and Their Applications
International Journal of Scientific & Engineering Research Volume 2, Issue 10, Oct-2011 1 ISSN 2229-5518 Survey of Databases Used in Image Processing and Their Applications Shubhpreet Kaur, Gagandeep Jindal Abstract- This paper gives review of Medical image database (MIDB) systems which have been developed in the past few years for research for medical fraternity and students. In this paper, I have surveyed all available medical image databases relevant for research and their use. Keywords: Image database, Medical Image Database System. —————————— —————————— 1. INTRODUCTION Measurement and recording techniques, such as electroencephalography, magnetoencephalography Medical imaging is the technique and process used to (MEG), Electrocardiography (EKG) and others, can create images of the human for clinical purposes be seen as forms of medical imaging. Image Analysis (medical procedures seeking to reveal, diagnose or is done to ensure database consistency and reliable examine disease) or medical science. As a discipline, image processing. it is part of biological imaging and incorporates radiology, nuclear medicine, investigative Open source software for medical image analysis radiological sciences, endoscopy, (medical) Several open source software packages are available thermography, medical photography and for performing analysis of medical images: microscopy. ImageJ 3D Slicer ITK Shubhpreet Kaur is currently pursuing masters degree OsiriX program in Computer Science and engineering in GemIdent Chandigarh Engineering College, Mohali, India. E-mail: MicroDicom [email protected] FreeSurfer Gagandeep Jindal is currently assistant processor in 1.1 Images used in Medical Research department Computer Science and Engineering in Here is the description of various modalities that are Chandigarh Engineering College, Mohali, India. E-mail: used for the purpose of research by medical and [email protected] engineering students as well as doctors. -

Face Recognition on a Smart Image Sensor Using Local Gradients
sensors Article Face Recognition on a Smart Image Sensor Using Local Gradients Wladimir Valenzuela 1 , Javier E. Soto 1, Payman Zarkesh-Ha 2 and Miguel Figueroa 1,* 1 Department of Electrical Engineering, Universidad de Concepción, Concepción 4070386, Chile; [email protected] (W.V.); [email protected] (J.E.S.) 2 Department of Electrical and Computer Engineering (ECE), University of New Mexico, Albuquerque, NM 87131-1070, USA; [email protected] * Correspondence: miguel.fi[email protected] Abstract: In this paper, we present the architecture of a smart imaging sensor (SIS) for face recognition, based on a custom-design smart pixel capable of computing local spatial gradients in the analog domain, and a digital coprocessor that performs image classification. The SIS uses spatial gradients to compute a lightweight version of local binary patterns (LBP), which we term ringed LBP (RLBP). Our face recognition method, which is based on Ahonen’s algorithm, operates in three stages: (1) it extracts local image features using RLBP, (2) it computes a feature vector using RLBP histograms, (3) it projects the vector onto a subspace that maximizes class separation and classifies the image using a nearest neighbor criterion. We designed the smart pixel using the TSMC 0.35 µm mixed- signal CMOS process, and evaluated its performance using postlayout parasitic extraction. We also designed and implemented the digital coprocessor on a Xilinx XC7Z020 field-programmable gate array. The smart pixel achieves a fill factor of 34% on the 0.35 µm process and 76% on a 0.18 µm process with 32 µm × 32 µm pixels. The pixel array operates at up to 556 frames per second. -
Image Analysis Image Analysis
PHYS871 Clinical Imaging ApplicaBons Image Analysis Image Analysis The Basics The Basics Dr Steve Barre April 2016 PHYS871 Clinical Imaging Applicaons / Image Analysis — The Basics 1 IntroducBon Image Processing Kernel Filters Rank Filters Fourier Filters Image Analysis Fourier Techniques Par4cle Analysis Specialist Solu4ons PHYS871 Clinical Imaging Applicaons / Image Analysis — The Basics 2 Image Display An image is a 2-dimensional collec4on of pixel values that can be displayed or printed by assigning a shade of grey (or colour) to every pixel value. PHYS871 Clinical Imaging Applicaons / Image Analysis — The Basics / Image Display 3 Image Display An image is a 2-dimensional collec4on of pixel values that can be displayed or printed by assigning a shade of grey (or colour) to every pixel value. PHYS871 Clinical Imaging Applicaons / Image Analysis — The Basics / Image Display 4 Spaal Calibraon For image analysis to produce meaningful results, the spaal calibraon of the image must be known. If the data acquisi4on parameters can be read from the image (or parameter) file then the spaal calibraon of the image can be determined. For simplicity and clarity, spaal calibraon will not be indicated on subsequent images. PHYS871 Clinical Imaging Applicaons / Image Analysis — The Basics / Image Display / Calibraon 5 SPM Image Display With Scanning Probe Microscope images there is no guarantee that the sample surface is level (so that the z values of the image are, on average, the same across the image). Raw image data A[er compensang for 4lt PHYS871 Clinical Imaging Applicaons / Image Analysis — The Basics / SPM Image Display 6 SPM Image Display By treang each scan line of an SPM image independently, anomalous jumps in the apparent height of the image (produced, for example, in STMs by abrupt changes in the tunnelling condi4ons) can be corrected for. -

Deep Learning in Medical Image Analysis
Journal of Imaging Editorial Deep Learning in Medical Image Analysis Yudong Zhang 1,* , Juan Manuel Gorriz 2 and Zhengchao Dong 3 1 School of Informatics, University of Leicester, Leicester LE1 7RH, UK 2 Department of Signal Theory, Telematics and Communications, University of Granada, 18071 Granada, Spain; [email protected] 3 Molecular Imaging and Neuropathology Division, Columbia University and New York State Psychiatric Institute, New York, NY 10032, USA; [email protected] * Correspondence: [email protected] Over recent years, deep learning (DL) has established itself as a powerful tool across a broad spectrum of domains in imaging—e.g., classification, prediction, detection, seg- mentation, diagnosis, interpretation, reconstruction, etc. While deep neural networks were initially nurtured in the computer vision community, they have quickly spread over medical imaging applications. The accelerating power of DL in diagnosing diseases will empower physicians and speed-up decision making in clinical environments. Applications of modern medical instru- ments and digitalization of medical care have led to enormous amounts of medical images being generated in recent years. In the big data arena, new DL methods and computational models for efficient data processing, analysis, and modeling of the generated data are crucial for clinical applications and understanding the underlying biological process. The purpose of this Special Issue (SI) “Deep Learning in Medical Image Analysis” is to present and highlight novel algorithms, architectures, techniques, and applications of DL for medical image analysis. This SI called for papers in April 2020. It received more than 60 submissions from Citation: Zhang, Y.; Gorriz, J.M.; over 30 different countries. -
PDF File, 212 KB
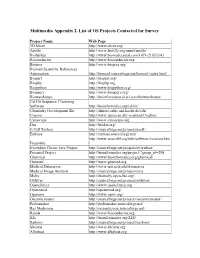
Multimedia Appendix 2. List of OS Projects Contacted for Survey Project Name Web Page 3D Slicer http://www.slicer.org/ Apollo http://www.fruitfly.org/annot/apollo/ Biobuilder http://www.biomedcentral.com/1471-2105/5/43 Bioconductor http://www.bioconductor.org Biojava http://www.biojava.org/ Biomail Scientific References Automation http://biomail.sourceforge.net/biomail/index.html Bioperl http://bioperl.org/ Biophp http://biophp.org Biopython http://www.biopython.org/ Bioquery http://www.bioquery.org/ Biowarehouse http://bioinformatics.ai.sri.com/biowarehouse/ Cd-Hit Sequence Clustering Software http://bioinformatics.org/cd-hit/ Chemistry Development Kit http://almost.cubic.uni-koeln.de/cdk/ Coasim http://www.daimi.au.dk/~mailund/CoaSim/ Cytoscape http://www.cytoscape.org Das http://biodas.org/ E-Cell System http://sourceforge.net/projects/ecell/ Emboss http://emboss.sourceforge.net/ http://www.ensembl.org/info/software/versions.htm Ensemble l Eviewbox Dicom Java Project http://sourceforge.net/projects/eviewbox/ Freemed Project http://bioinformatics.org/project/?group_id=298 Ghemical http://www.bioinformatics.org/ghemical/ Gnumed http://www.gnumed.org Medical Dataserver http://www.mii.ucla.edu/dataserver Medical Image Analysis http://sourceforge.net/projects/mia Moby http://biomoby.open-bio.org/ Olduvai http://sourceforge.net/projects/olduvai/ Openclinica http://www.openclinica.org Openemed http://openemed.org/ Openemr http://www.oemr.org/ Oscarmcmaster http://sourceforge.net/projects/oscarmcmaster/ Probemaker http://probemaker.sourceforge.net/