COPEIA February 1
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Carmine Shiner (Notropis Percobromus) in Canada
COSEWIC Assessment and Update Status Report on the Carmine Shiner Notropis percobromus in Canada THREATENED 2006 COSEWIC COSEPAC COMMITTEE ON THE STATUS OF COMITÉ SUR LA SITUATION ENDANGERED WILDLIFE DES ESPÈCES EN PÉRIL IN CANADA AU CANADA COSEWIC status reports are working documents used in assigning the status of wildlife species suspected of being at risk. This report may be cited as follows: COSEWIC 2006. COSEWIC assessment and update status report on the carmine shiner Notropis percobromus in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. vi + 29 pp. (www.sararegistry.gc.ca/status/status_e.cfm). Previous reports COSEWIC 2001. COSEWIC assessment and status report on the carmine shiner Notropis percobromus and rosyface shiner Notropis rubellus in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. v + 17 pp. Houston, J. 1994. COSEWIC status report on the rosyface shiner Notropis rubellus in Canada. Committee on the Status of Endangered Wildlife in Canada. Ottawa. 1-17 pp. Production note: COSEWIC would like to acknowledge D.B. Stewart for writing the update status report on the carmine shiner Notropis percobromus in Canada, prepared under contract with Environment Canada, overseen and edited by Robert Campbell, Co-chair, COSEWIC Freshwater Fishes Species Specialist Subcommittee. In 1994 and again in 2001, COSEWIC assessed minnows belonging to the rosyface shiner species complex, including those in Manitoba, as rosyface shiner (Notropis rubellus). For additional copies contact: COSEWIC Secretariat c/o Canadian Wildlife Service Environment Canada Ottawa, ON K1A 0H3 Tel.: (819) 997-4991 / (819) 953-3215 Fax: (819) 994-3684 E-mail: COSEWIC/[email protected] http://www.cosewic.gc.ca Également disponible en français sous le titre Évaluation et Rapport de situation du COSEPAC sur la tête carminée (Notropis percobromus) au Canada – Mise à jour. -

Annotated List of the Fishes of Illinois
STATE OF ILLINOIS HENRY HORNER' Gooernor DEPARTMENT OF REGISTRATION AND EDUCATION DIVISION OF THE NATURAL HISTORY SURVEY THEODORE H. FRISON, C,,'I Vol. XX BULLETIN Article V Annotated List of the Fishes of Illinois D. JOHN O'DONNELL PR,INTED BY AUTEORITY OF TEE STATE OF ILLINOIS URBANA, ILLINOIS AUGUST 1935 STATE OF ILLINOIS HrNrY HotNnn, Goz'ernor DEPARTMBNT OF REGISTRATION AND EDUCATION HoNonaslr .|oux J' Har"ltHaN, Director BOARD OF NATURAL RBSOURCES AND CONSERVATION HoNonasln JouN J. Halr,rHaN, Chairman Ph.D., LL'D., Chem.D', Wrr,r-rev Tnrlresr, D.Sc., LL.D., Biology Wrr,r,teu A. Novrs, D.Sc,, Chemistry Hunr.v C. Cowr,rs, Ph.D., D.Sc', Forestry EosoN C, Besnu, Ph.D., GeologY JonN W. At-vorn, C.8., Engineering Anrlun Currs WItlero, D.Eng., LL.D., Presid.ent ol the llnix,ersity of Illinois NATURAL HISTORY SURVEY DIVISION URBANA, ILLINOIS Scientifc and Technical 9taff Tnnoponn H. FrusoN, Ph.D., Chief BIOLOGY SECTION OF ECONOMIC ENTOMOLOGY SECTION OF AQUATIC Zoologist \4r. P. !'t-tsr, 8.5., Chiel Entomologist Devro H. TuoursoN, Ph.D., C. C. Covprot,M.S., Associate Entomologt.st F. D. Huwr, Field Naturalist Fo**o*, Ph. D:., Researe h Entomologist D. F. HeNsnN, A.M., Assistant Zoologist Vi. b. Zoologist S. C. CH,rxnirn, B.S., Southern Field Ento- D. J. O'DoNNnlr-, M.S., Assistant molo aist J. H. Bricen, 8.5., Central Field Entomolo- SECTION OF INSECT SURVEY qist Entomologisl L. Stt.o"snnB, M.5, Northetn Field En- H. H. Ross, Ph.D., Systematie [i. Entomolo' tomolostst C,rnr, O. -

Notropis Girardi) and Peppered Chub (Macrhybopsis Tetranema)
Arkansas River Shiner and Peppered Chub SSA, October 2018 Species Status Assessment Report for the Arkansas River Shiner (Notropis girardi) and Peppered Chub (Macrhybopsis tetranema) Arkansas River shiner (bottom left) and peppered chub (top right - two fish) (Photo credit U.S. Fish and Wildlife Service) Arkansas River Shiner and Peppered Chub SSA, October 2018 Version 1.0a October 2018 U.S. Fish and Wildlife Service Region 2 Albuquerque, NM This document was prepared by Angela Anders, Jennifer Smith-Castro, Peter Burck (U.S. Fish and Wildlife Service (USFWS) – Southwest Regional Office) Robert Allen, Debra Bills, Omar Bocanegra, Sean Edwards, Valerie Morgan (USFWS –Arlington, Texas Field Office), Ken Collins, Patricia Echo-Hawk, Daniel Fenner, Jonathan Fisher, Laurence Levesque, Jonna Polk (USFWS – Oklahoma Field Office), Stephen Davenport (USFWS – New Mexico Fish and Wildlife Conservation Office), Mark Horner, Susan Millsap (USFWS – New Mexico Field Office), Jonathan JaKa (USFWS – Headquarters), Jason Luginbill, and Vernon Tabor (Kansas Field Office). Suggested reference: U.S. Fish and Wildlife Service. 2018. Species status assessment report for the Arkansas River shiner (Notropis girardi) and peppered chub (Macrhybopsis tetranema), version 1.0, with appendices. October 2018. Albuquerque, NM. 172 pp. Arkansas River Shiner and Peppered Chub SSA, October 2018 EXECUTIVE SUMMARY ES.1 INTRODUCTION (CHAPTER 1) The Arkansas River shiner (Notropis girardi) and peppered chub (Macrhybopsis tetranema) are restricted primarily to the contiguous river segments of the South Canadian River basin spanning eastern New Mexico downstream to eastern Oklahoma (although the peppered chub is less widespread). Both species have experienced substantial declines in distribution and abundance due to habitat destruction and modification from stream dewatering or depletion from diversion of surface water and groundwater pumping, construction of impoundments, and water quality degradation. -

Redfin Shiner Lythrurus Umbratilis Kingdom: Animalia FEATURES Phylum: Chordata the Redfin Shiner Is About Two to Three Inches in Class: Osteichthyes Length
redfin shiner Lythrurus umbratilis Kingdom: Animalia FEATURES Phylum: Chordata The redfin shiner is about two to three inches in Class: Osteichthyes length. This minnow has a deep body that is Order: Cypriniformes compressed from side to side. The back is green- yellow with black specks, the sides are silver and the Family: Cyprinidae belly is white. There is a black spot at the front edge ILLINOIS STATUS (bottom) of the dorsal fin. The scales on the upper body are uniformly pigmented with black. The scales common, native in front of the dorsal fin are small and crowded. The lateral line curves sharply downward. Teeth are present in the throat. The breeding male has a bright blue body, red fins and white or pale gray on the top of the head. BEHAVIORS The redfin shiner may be found statewide in Illinois. It lives in clear, slow-flowing creeks. It is tolerant of © Konrad P. Schmidt, University of Minnesota silt and turbidity. This minnow is most common in pool habitats where there is little or no current. It adult swims in schools at middle water depths or near the surface. Spawning occurs from June through August over the nest of a green sunfish or longear sunfish. Approximately 500 to 700 eggs are deposited by the female. The redfin shiner eats insects and other small invertebrates. ILLINOIS RANGE © Illinois Department of Natural Resources. 2020. Biodiversity of Illinois. Unless otherwise noted, photos and images © Illinois Department of Natural Resources. © Uland Thomas Aquatic Habitats rivers and streams Woodland Habitats none Prairie and Edge Habitats none © Illinois Department of Natural Resources. -

RFP No. 212F for Endangered Species Research Projects for the Prairie Chub
1 RFP No. 212f for Endangered Species Research Projects for the Prairie Chub Final Report Contributing authors: David S. Ruppel, V. Alex Sotola, Ozlem Ablak Gurbuz, Noland H. Martin, and Timothy H. Bonner Addresses: Department of Biology, Texas State University, San Marcos, Texas 78666 (DSR, VAS, NHM, THB) Kirkkonaklar Anatolian High School, Turkish Ministry of Education, Ankara, Turkey (OAG) Principal investigators: Timothy H. Bonner and Noland H. Martin Email: [email protected], [email protected] Date: July 31, 2017 Style: American Fisheries Society Funding sources: Texas Comptroller of Public Accounts, Turkish Ministry of Education- Visiting Scholar Program (OAG) Summary Four hundred mesohabitats were sampled from 36 sites and 20 reaches within the upper Red River drainage from September 2015 through September 2016. Fishes (N = 36,211) taken from the mesohabitats represented 14 families and 49 species with the most abundant species consisting of Red Shiner Cyprinella lutrensis, Red River Shiner Notropis bairdi, Plains Minnow Hybognathus placitus, and Western Mosquitofish Gambusia affinis. Red River Pupfish Cyprinodon rubrofluviatilis (a species of greatest conservation need, SGCN) and Plains Killifish Fundulus zebrinus were more abundant within prairie streams (e.g., swift and shallow runs with sand and silt substrates) with high specific conductance. Red River Shiner (SGCN), Prairie Chub Macrhybopsis australis (SGCN), and Plains Minnow were more abundant within prairie 2 streams with lower specific conductance. The remaining 44 species of fishes were more abundant in non-prairie stream habitats with shallow to deep waters, which were more common in eastern tributaries of the upper Red River drainage and Red River mainstem. Prairie Chubs comprised 1.3% of the overall fish community and were most abundant in Pease River and Wichita River. -

Interim Performance Report Endangered Species
INTERIM PERFORMANCE REPORT ENDANGERED SPECIES PROGRAM GRANT NUMBER F17AP01052 WILDLIFE PROJECTS – ALABAMA PROJECT Reproductive Characteristics and Host Fish Determination of Canoe Creek Clubshell, Pleurobema athearni (Gangloff et al. 2006) in Big Canoe Creek drainage (Etowah and St. Clair Counties), Alabama October 1, 2018 - September 30, 2020 ALABAMA DEPARTMENT OF CONSERVATION AND NATURAL RESOURCES WILDLIFE AND FRESHWATER FISHERIES DIVISION Prepared by: Todd B. Fobian Alabama Division of Wildlife and Freshwater Fisheries PROJECT Reproductive Characteristics and Host Fish Determination of Canoe Creek Clubshell, Pleurobema athearni (Gangloff et al. 2006) in the Big Canoe Creek drainage (Etowah and St. Clair Counties), Alabama Year 1 Interim Report State: Alabama Introduction Pleurobema athearni (Gangloff et al, 2006), Canoe Creek Clubshell is currently a candidate for federally threatened/endangered status by U.S. Fish and Wildlife Service (FWS). It is Coosa Basin endemic, with historical records only known from the Big Canoe Creek (BCC) system in Alabama (Gangloff et al. 2006, Williams et al. 2008). Recent surveys completed by ADCNR and USFWS established the species is extant at six localities in the basin, with two in Upper Little Canoe Creek (ULCC), one in Lower Little Canoe Creek (LLCC), and three in BCC proper. (Fobian et al. 2017). As culture methods improve, propagated P. athearni juveniles could soon be available to support reintroduction/augmentation efforts within historical range. Little is known about Pleurobema athearni reproduction, female brooding period, or glochidial hosts. Female P. athearni are presumed short term-brooders and likely gravid from late spring to early summer (Gangloff et al. 2006, Williams et al. 2008). Glochidial hosts are currently unknown although other Mobile River Basin Pleurobema species often utilize Cyprinidae (shiners) to complete metamorphosis (Haag and Warren 1997, 2003, Weaver et al. -
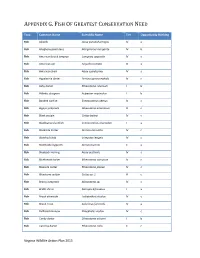
Fish of Greatest Conservation Need
APPENDIX G. FISH OF GREATEST CONSERVATION NEED Taxa Common Name Scientific Name Tier Opportunity Ranking Fish Alewife Alosa pseudoharengus IV a Fish Allegheny pearl dace Margariscus margarita IV b Fish American brook lamprey Lampetra appendix IV c Fish American eel Anguilla rostrata III a Fish American shad Alosa sapidissima IV a Fish Appalachia darter Percina gymnocephala IV c Fish Ashy darter Etheostoma cinereum I b Fish Atlantic sturgeon Acipenser oxyrinchus I b Fish Banded sunfish Enneacanthus obesus IV c Fish Bigeye jumprock Moxostoma ariommum III c Fish Black sculpin Cottus baileyi IV c Fish Blackbanded sunfish Enneacanthus chaetodon I a Fish Blackside darter Percina maculata IV c Fish Blotched chub Erimystax insignis IV c Fish Blotchside logperch Percina burtoni II a Fish Blueback Herring Alosa aestivalis IV a Fish Bluebreast darter Etheostoma camurum IV c Fish Blueside darter Etheostoma jessiae IV c Fish Bluestone sculpin Cottus sp. 1 III c Fish Brassy Jumprock Moxostoma sp. IV c Fish Bridle shiner Notropis bifrenatus I a Fish Brook silverside Labidesthes sicculus IV c Fish Brook Trout Salvelinus fontinalis IV a Fish Bullhead minnow Pimephales vigilax IV c Fish Candy darter Etheostoma osburni I b Fish Carolina darter Etheostoma collis II c Virginia Wildlife Action Plan 2015 APPENDIX G. FISH OF GREATEST CONSERVATION NEED Fish Carolina fantail darter Etheostoma brevispinum IV c Fish Channel darter Percina copelandi III c Fish Clinch dace Chrosomus sp. cf. saylori I a Fish Clinch sculpin Cottus sp. 4 III c Fish Dusky darter Percina sciera IV c Fish Duskytail darter Etheostoma percnurum I a Fish Emerald shiner Notropis atherinoides IV c Fish Fatlips minnow Phenacobius crassilabrum II c Fish Freshwater drum Aplodinotus grunniens III c Fish Golden Darter Etheostoma denoncourti II b Fish Greenfin darter Etheostoma chlorobranchium I b Fish Highback chub Hybopsis hypsinotus IV c Fish Highfin Shiner Notropis altipinnis IV c Fish Holston sculpin Cottus sp. -

Species Status Assessment (SSA) Report for the Ozark Chub (Erimystax Harryi) Version 1.2
Species Status Assessment (SSA) Report for the Ozark Chub (Erimystax harryi) Version 1.2 Ozark chub (Photo credit: Dustin Lynch, Arkansas Natural Heritage Commission) August 2019 U.S. Fish and Wildlife Service - Arkansas Ecological Services Field Office This document was prepared by Alyssa Bangs (U. S. Fish and Wildlife Service (USFWS) – Arkansas Ecological Services Field Office), Bryan Simmons (USFWS—Missouri Ecological Services Field Office), and Brian Evans (USFWS –Southeast Regional Office). We greatly appreciate the assistance of Jeff Quinn (Arkansas Game and Fish Commission), Brian Wagner (Arkansas Game and Fish Commission), and Jacob Westhoff (Missouri Department of Conservation) who provided helpful information and review of the draft document. We also thank the peer reviewers, who provided helpful comments. Suggested reference: U.S. Fish and Wildlife Service. 2019. Species status assessment report for the Ozark chub (Erimystax harryi). Version 1.2. August 2019. Atlanta, GA. CONTENTS Chapter 1: Executive Summary 1 1.1 Background 1 1.2 Analytical Framework 1 CHAPTER 2 – Species Information 4 2.1 Taxonomy and Genetics 4 2.2 Species Description 5 2.3 Range 6 Historical Range and Distribution 6 Current Range and Distribution 8 2.4 Life History Habitat 9 Growth and Longevity 9 Reproduction 9 Feeding 10 CHAPTER 3 –Factors Influencing Viability and Current Condition Analysis 12 3.1 Factors Influencing Viability 12 Sedimentation 12 Water Temperature and Flow 14 Impoundments 15 Water Chemistry 16 Habitat Fragmentation 17 3.2 Model 17 Analytical -

Endangered Species
FEATURE: ENDANGERED SPECIES Conservation Status of Imperiled North American Freshwater and Diadromous Fishes ABSTRACT: This is the third compilation of imperiled (i.e., endangered, threatened, vulnerable) plus extinct freshwater and diadromous fishes of North America prepared by the American Fisheries Society’s Endangered Species Committee. Since the last revision in 1989, imperilment of inland fishes has increased substantially. This list includes 700 extant taxa representing 133 genera and 36 families, a 92% increase over the 364 listed in 1989. The increase reflects the addition of distinct populations, previously non-imperiled fishes, and recently described or discovered taxa. Approximately 39% of described fish species of the continent are imperiled. There are 230 vulnerable, 190 threatened, and 280 endangered extant taxa, and 61 taxa presumed extinct or extirpated from nature. Of those that were imperiled in 1989, most (89%) are the same or worse in conservation status; only 6% have improved in status, and 5% were delisted for various reasons. Habitat degradation and nonindigenous species are the main threats to at-risk fishes, many of which are restricted to small ranges. Documenting the diversity and status of rare fishes is a critical step in identifying and implementing appropriate actions necessary for their protection and management. Howard L. Jelks, Frank McCormick, Stephen J. Walsh, Joseph S. Nelson, Noel M. Burkhead, Steven P. Platania, Salvador Contreras-Balderas, Brady A. Porter, Edmundo Díaz-Pardo, Claude B. Renaud, Dean A. Hendrickson, Juan Jacobo Schmitter-Soto, John Lyons, Eric B. Taylor, and Nicholas E. Mandrak, Melvin L. Warren, Jr. Jelks, Walsh, and Burkhead are research McCormick is a biologist with the biologists with the U.S. -

ECOLOGY of NORTH AMERICAN FRESHWATER FISHES
ECOLOGY of NORTH AMERICAN FRESHWATER FISHES Tables STEPHEN T. ROSS University of California Press Berkeley Los Angeles London © 2013 by The Regents of the University of California ISBN 978-0-520-24945-5 uucp-ross-book-color.indbcp-ross-book-color.indb 1 44/5/13/5/13 88:34:34 AAMM uucp-ross-book-color.indbcp-ross-book-color.indb 2 44/5/13/5/13 88:34:34 AAMM TABLE 1.1 Families Composing 95% of North American Freshwater Fish Species Ranked by the Number of Native Species Number Cumulative Family of species percent Cyprinidae 297 28 Percidae 186 45 Catostomidae 71 51 Poeciliidae 69 58 Ictaluridae 46 62 Goodeidae 45 66 Atherinopsidae 39 70 Salmonidae 38 74 Cyprinodontidae 35 77 Fundulidae 34 80 Centrarchidae 31 83 Cottidae 30 86 Petromyzontidae 21 88 Cichlidae 16 89 Clupeidae 10 90 Eleotridae 10 91 Acipenseridae 8 92 Osmeridae 6 92 Elassomatidae 6 93 Gobiidae 6 93 Amblyopsidae 6 94 Pimelodidae 6 94 Gasterosteidae 5 95 source: Compiled primarily from Mayden (1992), Nelson et al. (2004), and Miller and Norris (2005). uucp-ross-book-color.indbcp-ross-book-color.indb 3 44/5/13/5/13 88:34:34 AAMM TABLE 3.1 Biogeographic Relationships of Species from a Sample of Fishes from the Ouachita River, Arkansas, at the Confl uence with the Little Missouri River (Ross, pers. observ.) Origin/ Pre- Pleistocene Taxa distribution Source Highland Stoneroller, Campostoma spadiceum 2 Mayden 1987a; Blum et al. 2008; Cashner et al. 2010 Blacktail Shiner, Cyprinella venusta 3 Mayden 1987a Steelcolor Shiner, Cyprinella whipplei 1 Mayden 1987a Redfi n Shiner, Lythrurus umbratilis 4 Mayden 1987a Bigeye Shiner, Notropis boops 1 Wiley and Mayden 1985; Mayden 1987a Bullhead Minnow, Pimephales vigilax 4 Mayden 1987a Mountain Madtom, Noturus eleutherus 2a Mayden 1985, 1987a Creole Darter, Etheostoma collettei 2a Mayden 1985 Orangebelly Darter, Etheostoma radiosum 2a Page 1983; Mayden 1985, 1987a Speckled Darter, Etheostoma stigmaeum 3 Page 1983; Simon 1997 Redspot Darter, Etheostoma artesiae 3 Mayden 1985; Piller et al. -

Geological Survey of Alabama Calibration of The
GEOLOGICAL SURVEY OF ALABAMA Berry H. (Nick) Tew, Jr. State Geologist WATER INVESTIGATIONS PROGRAM CALIBRATION OF THE INDEX OF BIOTIC INTEGRITY FOR THE SOUTHERN PLAINS ICHTHYOREGION IN ALABAMA OPEN-FILE REPORT 0908 by Patrick E. O'Neil and Thomas E. Shepard Prepared in cooperation with the Alabama Department of Environmental Management and the Alabama Department of Conservation and Natural Resources Tuscaloosa, Alabama 2009 TABLE OF CONTENTS Abstract ............................................................ 1 Introduction.......................................................... 1 Acknowledgments .................................................... 6 Objectives........................................................... 7 Study area .......................................................... 7 Southern Plains ichthyoregion ...................................... 7 Methods ............................................................ 8 IBI sample collection ............................................. 8 Habitat measures............................................... 10 Habitat metrics ........................................... 12 The human disturbance gradient ................................... 15 IBI metrics and scoring criteria..................................... 19 Designation of guilds....................................... 20 Results and discussion................................................ 22 Sampling sites and collection results . 22 Selection and scoring of Southern Plains IBI metrics . 41 1. Number of native species ................................ -

Kansas Stream Fishes
A POCKET GUIDE TO Kansas Stream Fishes ■ ■ ■ ■ ■ ■ ■ ■ ■ ■ By Jessica Mounts Illustrations © Joseph Tomelleri Sponsored by Chickadee Checkoff, Westar Energy Green Team, Kansas Department of Wildlife, Parks and Tourism, Kansas Alliance for Wetlands & Streams, and Kansas Chapter of the American Fisheries Society Published by the Friends of the Great Plains Nature Center Table of Contents • Introduction • 2 • Fish Anatomy • 3 • Species Accounts: Sturgeons (Family Acipenseridae) • 4 ■ Shovelnose Sturgeon • 5 ■ Pallid Sturgeon • 6 Minnows (Family Cyprinidae) • 7 ■ Southern Redbelly Dace • 8 ■ Western Blacknose Dace • 9 ©Ryan Waters ■ Bluntface Shiner • 10 ■ Red Shiner • 10 ■ Spotfin Shiner • 11 ■ Central Stoneroller • 12 ■ Creek Chub • 12 ■ Peppered Chub / Shoal Chub • 13 Plains Minnow ■ Silver Chub • 14 ■ Hornyhead Chub / Redspot Chub • 15 ■ Gravel Chub • 16 ■ Brassy Minnow • 17 ■ Plains Minnow / Western Silvery Minnow • 18 ■ Cardinal Shiner • 19 ■ Common Shiner • 20 ■ Bigmouth Shiner • 21 ■ • 21 Redfin Shiner Cover Photo: Photo by Ryan ■ Carmine Shiner • 22 Waters. KDWPT Stream ■ Golden Shiner • 22 Survey and Assessment ■ Program collected these Topeka Shiner • 23 male Orangespotted Sunfish ■ Bluntnose Minnow • 24 from Buckner Creek in Hodgeman County, Kansas. ■ Bigeye Shiner • 25 The fish were catalogued ■ Emerald Shiner • 26 and returned to the stream ■ Sand Shiner • 26 after the photograph. ■ Bullhead Minnow • 27 ■ Fathead Minnow • 27 ■ Slim Minnow • 28 ■ Suckermouth Minnow • 28 Suckers (Family Catostomidae) • 29 ■ River Carpsucker •