Tunicates Push the Limits of Animal Evo-Devo David EK Ferrier*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
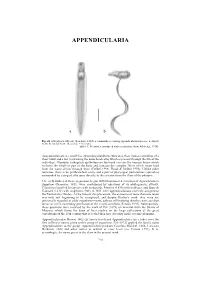
Appendicularia of CTAW
APPENDICULARIA APPENDICULARIA a b Fig. 26. Oikopleura albicans (Leuckart, 1854), a commonly occurring appendicularian species: a, dorsal view; b, lateral view. (Scale bar = 0.1 mm). [after T. Prentiss, reproduced with permission, from Alldredge 1976] Appendicularians are small free swimming planktonic tunicates, their bodies consisting of a short trunk and a tail (containing the notochord cells) which is present through the life of the individual. Glandular (oikoplast) epithelium on the trunk secretes the mucous house which encloses the whole or part of the body and contains the complex filters which strain food from the water driven through them (Deibel 1998; Flood & Deibel 1998). Unlike other tunicates, there is no peribranchial cavity and a pair of pharyngeal perforations (spiracles) surrounded by a ring of cilia open directly to the exterior from the floor of the pharynx. The early studies of these organisms, begun with Chamisso's description of Appendicularia flagellum Chamisso, 1821, were confounded by questions of its phylogenetic affinity. Chamisso classified his species with medusoids, Mertens (1830) with molluscs, and Quoy & Gaimard (1833) with zoophytes. Only in 1851 were appendicularians correctly assigned to the Tunicata by Huxley. At the time of this placement, the existence of more than one taxon was only just beginning to be recognised, and despite Huxley's work, they were not universally regarded as adult organisms—some authors still insisting that they were ascidian larvae or a free swimming generation of the sessile ascidians (Fenaux 1993). Subsequently, these questions were resolved by the work of Fol (1872) on material from the Straits of Messina, which forms the basis of later studies on the large collections of the great expeditions of the 19th century that revealed their true diversity in the oceanic plankton. -

Brains of Primitive Chordates 439
Brains of Primitive Chordates 439 Brains of Primitive Chordates J C Glover, University of Oslo, Oslo, Norway although providing a more direct link to the evolu- B Fritzsch, Creighton University, Omaha, NE, USA tionary clock, is nevertheless hampered by differing ã 2009 Elsevier Ltd. All rights reserved. rates of evolution, both among species and among genes, and a still largely deficient fossil record. Until recently, it was widely accepted, both on morpholog- ical and molecular grounds, that cephalochordates Introduction and craniates were sister taxons, with urochordates Craniates (which include the sister taxa vertebrata being more distant craniate relatives and with hemi- and hyperotreti, or hagfishes) represent the most chordates being more closely related to echinoderms complex organisms in the chordate phylum, particu- (Figure 1(a)). The molecular data only weakly sup- larly with respect to the organization and function of ported a coherent chordate taxon, however, indicat- the central nervous system. How brain complexity ing that apparent morphological similarities among has arisen during evolution is one of the most chordates are imposed on deep divisions among the fascinating questions facing modern science, and it extant deuterostome taxa. Recent analysis of a sub- speaks directly to the more philosophical question stantially larger number of genes has reversed the of what makes us human. Considerable interest has positions of cephalochordates and urochordates, pro- therefore been directed toward understanding the moting the latter to the most closely related craniate genetic and developmental underpinnings of nervous relatives (Figure 1(b)). system organization in our more ‘primitive’ chordate relatives, in the search for the origins of the vertebrate Comparative Appearance of Brains, brain in a common chordate ancestor. -

Spatial Distribution Patterns of Appendicularians in the Drake Passage: Potential Indicators of Water Masses?
diversity Article Spatial Distribution Patterns of Appendicularians in the Drake Passage: Potential Indicators of Water Masses? Marcin Kalarus 1 and Anna Panasiuk 2,* 1 Department of Aquatic Ecology, Maritime Institute, Gdynia Maritime University, Smoluchowskiego 1/3, 80-214 Gda´nsk,Poland; [email protected] 2 Department of Marine Plankton Research, Faculty of Oceanography and Geography, Institute of Oceanography, University of Gda´nsk,Av. Marszałka J. Piłsudskiego 46, 81-378 Gdynia, Poland * Correspondence: [email protected]; Tel.: +485-8523-6844 Abstract: Appendicularians are one of the most common animals found within zooplankton as- semblages. They play a very important role as filter feeders but are, unfortunately, inconsistently reported in the Antarctic literature. The present paper attempts to describe the zonal diversity of appendicularians and the main environmental factors influencing their communities in the Drake Passage. Samples were collected during Antarctic summer in 2009–2010. A total of eight species of larvaceans were identified. Fritillaria borealis was the species found in the highest numbers in almost the entire studied area, and was observed at all sampling stations. The distributions of other taxa were limited to specific hydrological zones and hydrological conditions. F. fraudax and Oikopleura gaussica were typical of the areas between the Polar Front and the Subantarctic Front zones, and their distributions were significantly correlated with temperature and salinity, likely making them good indicator species. The F. fusiformis distribution was strictly related to South American waters. In summary, temperature was the strongest environmental factor influencing the larvacean community structure in the Drake Passage, and we also found that testing environmental factors on larvaceans Citation: Kalarus, M.; Panasiuk, A. -

A Chordate Species Lacking Nodal Utilizes Calcium Oscillation and Bmp for Left–Right Patterning
A chordate species lacking Nodal utilizes calcium oscillation and Bmp for left–right patterning Takeshi A. Onumaa,1, Momoko Hayashia, Fuki Gyojab, Kanae Kishia, Kai Wanga, and Hiroki Nishidaa aDepartment of Biological Sciences, Graduate School of Science, Osaka University, 560-0043 Toyonaka, Osaka, Japan; and bMarine Genomics Unit, Okinawa Institute of Science and Technology Graduate University, 904-0495 Onna, Okinawa, Japan Edited by Edward M. De Robertis, David Geffen School of Medicine, University of California, Los Angeles, CA, and approved December 20, 2019 (received for review September 27, 2019) Larvaceans are chordates with a tadpole-like morphology. In neurula by ciliary movement (27, 28). These are considered to contrast to most chordates of which early embryonic morphology is be biological cues that induce the left-sided expression of Nodal,a bilaterally symmetric and the left–right (L–R) axis is specified by the member of the TGF-β family, leading to the L–R asymmetry of Nodal pathway later on, invariant L–R asymmetry emerges in four- Deuterostomia (e.g., vertebrates) and Lophotrochozoa (e.g., pond cell embryos of larvaceans. The asymmetric cell arrangements exist snails). However, these mechanisms are not universal. For ex- through development of the tailbud. The tail thus twists 90° in a ample, motile cilia are not associated with L–R patterning in the counterclockwise direction relative to the trunk, and the tail nerve pond snail (29, 30), and Nodal is lost in Ecdysozoa (e.g., nem- cord localizes on the left side. Here, we demonstrate that larvacean atodes and flies). This diversity of symmetry-breaking mecha- embryos have nonconventional L–R asymmetries: 1) L- and R-cells of nisms suggests that there may be organisms with novel strategies 2+ the two-cell embryo had remarkably asymmetric cell fates; 2) Ca for L–R patterning. -

Ascidian News #82 December 2018
ASCIDIAN NEWS* Gretchen Lambert 12001 11th Ave. NW, Seattle, WA 98177 206-365-3734 [email protected] home page: http://depts.washington.edu/ascidian/ Number 82 December 2018 A big thank-you to all who sent in contributions. There are 85 New Publications listed at the end of this issue. Please continue to send me articles, and your new papers, to be included in the June 2019 issue of AN. It’s never too soon to plan ahead. *Ascidian News is not part of the scientific literature and should not be cited as such. NEWS AND VIEWS 1. From Stefano Tiozzo ([email protected]) and Remi Dumollard ([email protected]): The 10th Intl. Tunicata Meeting will be held at the citadel of Saint Helme in Villefranche sur Mer (France), 8- 12 July 2019. The web site with all the information will be soon available, save the date! We are looking forward to seeing you here in the Riviera. A bientôt! Remi and Stefano 2. The 10th Intl. Conference on Marine Bioinvasions was held in Puerto Madryn, Patagonia, Argentina, October 16-18. At the conference website (http://www.marinebioinvasions.info/index) the program and abstracts in pdf can be downloaded. Dr. Rosana Rocha presented one of the keynote talks: "Ascidians in the anthropocene - invasions waiting to happen". See below under Meetings Abstracts for all the ascidian abstracts; my thanks to Evangelina Schwindt for compiling them. The next (11th) meeting will be in Annapolis, Maryland, organized by Greg Ruiz, Smithsonian Invasions lab, date to be determined. 3. Conference proceedings of the May 2018 Invasive Sea Squirt Conference will be peer reviewed and published in a special issue of the REABIC journal Management of Biological Invasions. -

Oikopleura Dioica
To: [email protected] o ABSTRACT We propose that a BAC library should be constructed for the larvacean urochordate Oikopleura dioica. This animal may be the most similar among extant animals to the last common ancestor of all chordates, and thus, its investigation can illuminate the mechanisms causing the vertebrate genome expansion, the principles that guide the evolution of functions of duplicate genes in development, and the origin of vertebrate developmental innovations. Oikopleura dioica has a genome of about 70 Mb, the smallest of any chordate. o IMPORTANCE OF THE ORGANISM TO BIOMEDICAL OR BIOLOGICAL RESEARCH. The Animal. Oikopleura dioica is a 3 mm long urochordate, an abundant filter-feeder in marine plankton. Oikopleura and other members of its phylogenetic class, the larvaceans (or appendicularians) remain free swimming and tadpole-like into sexual maturity. Because larvaceans are basally diverging among the urochordates, this free-swimming life style may have been ancestral (Wada 1998). Larvaceans have more functional organ systems than do ascidian larvae; their embryos, tadpole larvae, and adults are transparent, which facilitates observations on all aspects of development (Fig. 1); embryonic development takes only 14 hours; and the generation time is less than two weeks. Furthermore, Oikopleura dioica is readily available from plankton tows for much of the year off Northern hemisphere coasts. Oikopleura dioica can be among the most abundant animals in offshore waters, even more than copepods, amounting to several animals per liter (Dagg & Ortner 1994; Capitano & Esnal 1998). With Oikopleura, one can perform single-pair matings, and culture the offspring in the lab continuously in the presence of fresh sea water (Fenaux & Gorsky, 1983; Bassham, unpubl). -

Telomere-To-Telomere Assembly of the Genome of an Individual Oikopleura
Bliznina et al. BMC Genomics (2021) 22:222 https://doi.org/10.1186/s12864-021-07512-6 RESEARCH ARTICLE Open Access Telomere-to-telomere assembly of the genome of an individual Oikopleura dioica from Okinawa using Nanopore-based sequencing Aleksandra Bliznina1* , Aki Masunaga1, Michael J. Mansfield1, Yongkai Tan1, Andrew W. Liu1, Charlotte West1,2, Tanmay Rustagi1, Hsiao-Chiao Chien1, Saurabh Kumar1, Julien Pichon1, Charles Plessy1* and Nicholas M. Luscombe1,2,3 Abstract Background: The larvacean Oikopleura dioica is an abundant tunicate plankton with the smallest (65–70 Mbp) non- parasitic, non-extremophile animal genome identified to date. Currently, there are two genomes available for the Bergen (OdB3) and Osaka (OSKA2016) O. dioica laboratory strains. Both assemblies have full genome coverage and high sequence accuracy. However, a chromosome-scale assembly has not yet been achieved. Results: Here, we present a chromosome-scale genome assembly (OKI2018_I69) of the Okinawan O. dioica produced using long-read Nanopore and short-read Illumina sequencing data from a single male, combined with Hi-C chromosomal conformation capture data for scaffolding. The OKI2018_I69 assembly has a total length of 64.3 Mbp distributed among 19 scaffolds. 99% of the assembly is contained within five megabase-scale scaffolds. We found telomeres on both ends of the two largest scaffolds, which represent assemblies of two fully contiguous autosomal chromosomes. Each of the other three large scaffolds have telomeres at one end only and we propose that they correspond to sex chromosomes split into a pseudo-autosomal region and X-specific or Y-specific regions. Indeed, these five scaffolds mostly correspond to equivalent linkage groups in OdB3, suggesting overall agreement in chromosomal organization between the two populations. -

Evolutionary Diversification of Secondary Mechanoreceptor Cells
Rigon et al. BMC Evolutionary Biology 2013, 13:112 http://www.biomedcentral.com/1471-2148/13/112 RESEARCH ARTICLE Open Access Evolutionary diversification of secondary mechanoreceptor cells in tunicata Francesca Rigon1, Thomas Stach2, Federico Caicci1, Fabio Gasparini1*, Paolo Burighel1 and Lucia Manni1 Abstract Background: Hair cells are vertebrate secondary sensory cells located in the ear and in the lateral line organ. Until recently, these cells were considered to be mechanoreceptors exclusively found in vertebrates that evolved within this group. Evidence of secondary mechanoreceptors in some tunicates, the proposed sister group of vertebrates, has recently led to the hypothesis that vertebrate and tunicate secondary sensory cells share a common origin. Secondary sensory cells were described in detail in two tunicate groups, ascidians and thaliaceans, in which they constitute an oral sensory structure called the coronal organ. Among thaliaceans, the organ is absent in salps and it has been hypothesised that this condition is due to a different feeding system adopted by this group of animals. No information is available as to whether a comparable structure exists in the third group of tunicates, the appendicularians, although different sensory structures are known to be present in these animals. Results: We studied the detailed morphology of appendicularian oral mechanoreceptors. Using light and electron microscopy we could demonstrate that the mechanosensory organ called the circumoral ring is composed of secondary sensory cells. We described the ultrastructure of the circumoral organ in two appendicularian species, Oikopleura dioica and Oikopleura albicans, and thus taxonomically completed the data collection of tunicate secondary sensory cells. To understand the evolution of secondary sensory cells in tunicates, we performed a cladistic analysis using morphological data. -

Ascidian News #76 December 2015
ASCIDIAN NEWS* Gretchen Lambert 12001 11th Ave. NW, Seattle, WA 98177 206-365-3734 [email protected] home page: http://depts.washington.edu/ascidian/ Number 76 December 2015 I greatly enjoyed participating in teaching a two week ascidian course at Nagoya University’s Sugashima Marine Lab from the end of June to July 10, and then attended the Intl. Tunicata meeting in Aomori, Japan from July 13-17. This issue is the second for my 40th year of compiling Ascidian News. I would greatly appreciate hearing from you whether you still find it useful and interesting. There are 93 New Publications listed at the end of this issue. *Ascidian News is not part of the scientific literature and should not be cited as such. NEWS AND VIEWS 1. Ciona intestinalis now shown to be 2 separate species. Because so many researchers work on Ciona intestinalis, and so many papers are published on this species, I draw your attention to 2 new publications showing at last that Ciona intestinalis A and B are different species and designating the correct names to be used in all future publications: Brunetti, R., Gissi, C., Pennati, R., Caicci, F., Gasparini, F. and Manni, L. 2015. Morphological evidence indicates that Ciona intestinalis (Tunicata, Ascidiacea) type A and type B are different species. Journal of Zool. Systematics & Evolutionary Research 53 (3): 186–193. [Type A is now designated C. robusta; type B retains the name C. intestinalis.] The second new publication describes larval differences between the two species: Pennati, R., Ficetola, G. F., Brunetti, R., Caicci, F., Gasparini, F., Griggio, F., Sato, A., Stach, T., Kaul-Strehlow, S., Gissi, C. -

Long-Distance Cell Migration During Larval Development in the Appendicularian, Oikopleura Dioica
Developmental Biology 395 (2014) 299–306 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/locate/developmentalbiology Long-distance cell migration during larval development in the appendicularian, Oikopleura dioica Kanae Kishi n, Takeshi A. Onuma, Hiroki Nishida Department of Biological Sciences, Graduate School of Science, Osaka University, 1-1 Machikaneyama-cho, Toyonaka 560-0043, Osaka, Japan article info abstract Article history: The appendicularian, Oikopleura dioica, is a planktonic chordate. Its simple and transparent body, Received 18 July 2014 invariant cell lineages and short life cycle of 5 days make it a promising model organism for studies of Received in revised form chordate development. Here we describe the cell migration that occurs during development of the O. 30 August 2014 dioica larva. Using time-lapse imaging facilitated by florescent labeling of cells, three cell populations Accepted 7 September 2014 exhibiting long-distance migration were identified and characterized. These included (i) a multi- Available online 16 September 2014 nucleated oral gland precursor that migrates anteriorly within the trunk region and eventually separates Keywords: into the left and right sides, (ii) endodermal strand cells that are collectively retracted from the tail into Cell migration the trunk in a tractor movement, and (iii) two subchordal cell precursors that individually migrate out Chordate from the trunk to the tip of the tail. The migration of subchordal cell precursors starts when all of the Appendicularian endodermal strand cells enter the trunk, and follows the same path but in a direction opposite to that of Oikopleura dioica fl Chemoattractant the latter. Labeling of these cells with a photoconvertible uorescent protein, Kaede, demonstrated that Endodermal strand the endodermal strand cells and subchordal cell precursors have distinct origins and eventual fates. -

Phylogenetic Analysis of Phenotypic Characters of Tunicata Supports Basal Appendicularia and Monophyletic Ascidiacea
Cladistics Cladistics 36 (2020) 259–300 10.1111/cla.12405 Phylogenetic analysis of phenotypic characters of Tunicata supports basal Appendicularia and monophyletic Ascidiacea Katrin Brauna, Fanny Leubnerb and Thomas Stachc,* aVergleichende Zoologie, Institut fur€ Biologie, Humboldt-Universitat€ zu Berlin, Philippstrasse 13, Haus 2, 10115 Berlin, Germany; bAnimal Evolution and Biodiversity, J-F-Blumenbach Institute for Zoology & Anthropology, Georg-August-University Gottingen,€ Untere Karspule€ 2, 37073 Gottingen,€ Germany; cMolekulare Parasitologie, Institut fur€ Biologie, Humboldt-Universitat€ zu Berlin, Philippstrasse 13, Haus 14, 10115 Berlin, Germany Abstract With approximately 3000 marine species, Tunicata represents the most disparate subtaxon of Chordata. Molecular phyloge- netic studies support Tunicata as sister taxon to Craniota, rendering it pivotal to understanding craniate evolution. Although successively more molecular data have become available to resolve internal tunicate phylogenetic relationships, phenotypic data have not been utilized consistently. Herein these shortcomings are addressed by cladistically analyzing 117 phenotypic characters for 49 tunicate species comprising all higher tunicate taxa, and five craniate and cephalochordate outgroup species. In addition, a combined analysis of the phenotypic characters with 18S rDNA-sequence data is performed in 32 OTUs. The analysis of the combined data is congruent with published molecular analyses. Successively up-weighting phenotypic characters indicates that phenotypic data contribute disproportionally more to the resulting phylogenetic hypothesis. The strict consensus tree from the analysis of the phenotypic characters as well as the single most parsimonious tree found in the analysis of the combined dataset recover monophyletic Appendicularia as sister taxon to the remaining tunicate taxa. Thus, both datasets support the hypothesis that the last common ancestor of Tunicata was free-living and that ascidian sessility is a derived trait within Tunicata. -

Hox Cluster Disintegration with Persistent Anteroposterior Order Of
letters to nature 22. Pearce, J. M. in Animal Learning and Cognition (ed. Mackintosh, N. J.) 110–134 (Academic, San Diego, Hox genes are involved in establishing morphological identities 1994). along the anteroposterior axis of bilaterians and cnidarians2. Phylo- 23. Fisher, R. A. The Genetical Theory of Natural Selection (Oxford Univ. Press, Oxford, 1930). 24. Glanville, P. W. & Allen, J. A. Protective polymorphism in populations of computer-simulated moth- genetic analysis suggests that the ancestor of all bilaterians had at like prey. Oikos 80, 565–571 (1997). least seven Hox genes—five anterior, one central and one posterior, 25. Go¨tmark, F. & Hohlfa¨lt, A. Bright male plumage and predation risk in passerine birds: are males easier according to the nomenclature of ref. 3—grouped in a genomic to detect than females? Oikos 74, 475–484 (1995). 26. Sherratt, T. N. & Beatty, C. D. The evolution of warning signals as reliable indicators of prey defense. cluster where the gene order correlated with sequential expression Am. Nat. 162, 377–389 (2003). along the anteroposterior axis. Both Hox gene sequences and the 27. Miller, G. A. The magical number seven, plus or minus two: some limits on our capacity for processing Hox cluster evolved in distinct animal lineages, with occasional information. Psychol. Rev. 63, 81–97 (1956). cluster splits in invertebrate protostomes (Drosophila, Caenorhab- 28. Dukas, R. Behavioural and ecological consequences of limited attention. Philos. Trans. R. Soc. Lond. B 4,5 357, 1539–1548 (2002). ditis), and gene losses . The significance of these discrete alterations 29. Beatty, C. D., Bain, R.