3D Reconstruction of Structures of Hatched Larva and Young Juvenile Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

National Monitoring Program for Biodiversity and Non-Indigenous Species in Egypt
UNITED NATIONS ENVIRONMENT PROGRAM MEDITERRANEAN ACTION PLAN REGIONAL ACTIVITY CENTRE FOR SPECIALLY PROTECTED AREAS National monitoring program for biodiversity and non-indigenous species in Egypt PROF. MOUSTAFA M. FOUDA April 2017 1 Study required and financed by: Regional Activity Centre for Specially Protected Areas Boulevard du Leader Yasser Arafat BP 337 1080 Tunis Cedex – Tunisie Responsible of the study: Mehdi Aissi, EcApMEDII Programme officer In charge of the study: Prof. Moustafa M. Fouda Mr. Mohamed Said Abdelwarith Mr. Mahmoud Fawzy Kamel Ministry of Environment, Egyptian Environmental Affairs Agency (EEAA) With the participation of: Name, qualification and original institution of all the participants in the study (field mission or participation of national institutions) 2 TABLE OF CONTENTS page Acknowledgements 4 Preamble 5 Chapter 1: Introduction 9 Chapter 2: Institutional and regulatory aspects 40 Chapter 3: Scientific Aspects 49 Chapter 4: Development of monitoring program 59 Chapter 5: Existing Monitoring Program in Egypt 91 1. Monitoring program for habitat mapping 103 2. Marine MAMMALS monitoring program 109 3. Marine Turtles Monitoring Program 115 4. Monitoring Program for Seabirds 118 5. Non-Indigenous Species Monitoring Program 123 Chapter 6: Implementation / Operational Plan 131 Selected References 133 Annexes 143 3 AKNOWLEGEMENTS We would like to thank RAC/ SPA and EU for providing financial and technical assistances to prepare this monitoring programme. The preparation of this programme was the result of several contacts and interviews with many stakeholders from Government, research institutions, NGOs and fishermen. The author would like to express thanks to all for their support. In addition; we would like to acknowledge all participants who attended the workshop and represented the following institutions: 1. -
Appendicularia of CTAW
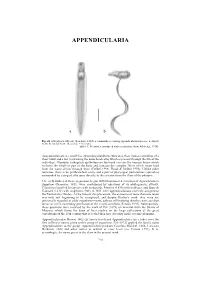
APPENDICULARIA APPENDICULARIA a b Fig. 26. Oikopleura albicans (Leuckart, 1854), a commonly occurring appendicularian species: a, dorsal view; b, lateral view. (Scale bar = 0.1 mm). [after T. Prentiss, reproduced with permission, from Alldredge 1976] Appendicularians are small free swimming planktonic tunicates, their bodies consisting of a short trunk and a tail (containing the notochord cells) which is present through the life of the individual. Glandular (oikoplast) epithelium on the trunk secretes the mucous house which encloses the whole or part of the body and contains the complex filters which strain food from the water driven through them (Deibel 1998; Flood & Deibel 1998). Unlike other tunicates, there is no peribranchial cavity and a pair of pharyngeal perforations (spiracles) surrounded by a ring of cilia open directly to the exterior from the floor of the pharynx. The early studies of these organisms, begun with Chamisso's description of Appendicularia flagellum Chamisso, 1821, were confounded by questions of its phylogenetic affinity. Chamisso classified his species with medusoids, Mertens (1830) with molluscs, and Quoy & Gaimard (1833) with zoophytes. Only in 1851 were appendicularians correctly assigned to the Tunicata by Huxley. At the time of this placement, the existence of more than one taxon was only just beginning to be recognised, and despite Huxley's work, they were not universally regarded as adult organisms—some authors still insisting that they were ascidian larvae or a free swimming generation of the sessile ascidians (Fenaux 1993). Subsequently, these questions were resolved by the work of Fol (1872) on material from the Straits of Messina, which forms the basis of later studies on the large collections of the great expeditions of the 19th century that revealed their true diversity in the oceanic plankton. -

Canestro 07Devbio Oikopleura Retinoic Acid Chordate.Pdf
Developmental Biology 305 (2007) 522–538 www.elsevier.com/locate/ydbio Development of a chordate anterior–posterior axis without classical retinoic acid signaling ⁎ Cristian Cañestro, John H. Postlethwait Institute of Neuroscience, University of Oregon, Eugene, OR 97403, USA Received for publication 28 November 2006; revised 18 February 2007; accepted 26 February 2007 Available online 2 March 2007 Abstract Developmental signaling by retinoic acid (RA) is thought to be an innovation essential for the origin of the chordate body plan. The larvacean urochordate Oikopleura dioica maintains a chordate body plan throughout life, and yet its genome appears to lack genes for RA synthesis, degradation, and reception. This suggests the hypothesis that the RA-machinery was lost during larvacean evolution, and predicts that Oikopleura development has become independent of RA-signaling. This prediction raises the problem that the anterior–posterior organization of a chordate body plan can be developed without the classical morphogenetic role of RA. To address this problem, we performed pharmacological treatments and analyses of developmental molecular markers to investigate whether RA acts in anterior–posterior axial patterning in Oikopleura embryos. Results revealed that RA does not cause homeotic posteriorization in Oikopleura as it does in vertebrates and cephalochordates, and showed that a chordate can develop the phylotypic body plan in the absence of the classical morphogenetic role of RA. A comparison of Oikopleura and ascidian evidence suggests that the lack of RA-induced homeotic posteriorization is a shared derived feature of urochordates. We discuss possible relationships of altered roles of RA in urochordate development to genomic events, such as rupture of the Hox-cluster, in the context of a new understanding of chordate phylogeny. -

Study of Dental Fluorosis in Subjects Related to a Phosphatic Fertilizer
Indian Journal of Geo Marine Sciences Vol. 46 (07), July 2017, pp. 1371-1380 Diversity and abundance of epipelagic larvaceans and calanoid copepods in the eastern equatorial Indian Ocean during the spring inter-monsoon Kaizhi Li, Jianqiang Yin*, Yehui Tan, Liangmin Huang & Gang Li Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China *[E-mail: [email protected]] Received 20 August 2015; revised 29 September 2015 This study investigated the species composition, distribution and abundance of larvaceans and calanoid copepods in the eastern equatorial Indian Ocean. In total, 25 species of larvaceans and 69 species of calanoid copepods were identified in the study area. Although the average diversity and evenness indexes of larvaceans were lower than those of calanoid copepods, the abundance of larvaceans was higher than that of calanoids with means of 40.1±14.9 ind m-3 and 28.4±9.1 ind m-3, respectively. Larvacean community was numerically dominated by Oikopleura fusiformis, Oikopleura longicauda, Oikopleura cophocerca, Fritillaria formica and Fritillaria pellucida, accounting for 83% of total larvacean abundance. The calanoid community was dominated by the following five species, represented 61% of calanoid copepods: Clausocalanus furcatus, Clausocalanus farrani, Acartia negligens, Acrocalanus longicornis as well as the copepodite stage of Euchaeta spp. This study highlights that the importance of larvaceans in the eastern equatorial Indian Ocean. [Keywords: Appendicularians, Calanoids, Water mass, Monsoon, Indian Ocean] Introduction It has been clear that small organisms of marine Clausocalanus) and the cyclopoid genera (such as zooplankton have historically been under-sampled by Oithona, Oncaea and Corycaeus1). -

Brains of Primitive Chordates 439
Brains of Primitive Chordates 439 Brains of Primitive Chordates J C Glover, University of Oslo, Oslo, Norway although providing a more direct link to the evolu- B Fritzsch, Creighton University, Omaha, NE, USA tionary clock, is nevertheless hampered by differing ã 2009 Elsevier Ltd. All rights reserved. rates of evolution, both among species and among genes, and a still largely deficient fossil record. Until recently, it was widely accepted, both on morpholog- ical and molecular grounds, that cephalochordates Introduction and craniates were sister taxons, with urochordates Craniates (which include the sister taxa vertebrata being more distant craniate relatives and with hemi- and hyperotreti, or hagfishes) represent the most chordates being more closely related to echinoderms complex organisms in the chordate phylum, particu- (Figure 1(a)). The molecular data only weakly sup- larly with respect to the organization and function of ported a coherent chordate taxon, however, indicat- the central nervous system. How brain complexity ing that apparent morphological similarities among has arisen during evolution is one of the most chordates are imposed on deep divisions among the fascinating questions facing modern science, and it extant deuterostome taxa. Recent analysis of a sub- speaks directly to the more philosophical question stantially larger number of genes has reversed the of what makes us human. Considerable interest has positions of cephalochordates and urochordates, pro- therefore been directed toward understanding the moting the latter to the most closely related craniate genetic and developmental underpinnings of nervous relatives (Figure 1(b)). system organization in our more ‘primitive’ chordate relatives, in the search for the origins of the vertebrate Comparative Appearance of Brains, brain in a common chordate ancestor. -

Science Journals
SCIENCE ADVANCES | RESEARCH ARTICLE MARINE ECOLOGY Copyright © 2017 The Authors, some From the surface to the seafloor: How giant larvaceans rights reserved; exclusive licensee transport microplastics into the deep sea American Association for the Advancement † † of Science. No claim to Kakani Katija,* C. Anela Choy,* Rob E. Sherlock, Alana D. Sherman, Bruce H. Robison original U.S. Government Works. Distributed Plastic waste is a pervasive feature of marine environments, yet little is empirically known about the biological under a Creative and physical processes that transport plastics through marine ecosystems. To address this need, we conducted Commons Attribution in situ feeding studies of microplastic particles (10 to 600 mm in diameter) with the giant larvacean Bathochordaeus NonCommercial stygius. Larvaceans are abundant components of global zooplankton assemblages, regularly build mucus “houses” License 4.0 (CC BY-NC). to filter particulate matter from the surrounding water, and later abandon these structures when clogged. By conducting in situ feeding experiments with remotely operated vehicles, we show that giant larvaceans are able to filter a range of microplastic particles from the water column, ingest, and then package microplastics into their fecal pellets. Microplastics also readily affix to their houses, which have been shown to sink quickly to the seafloor and deliver pulses of carbon to benthic ecosystems. Thus, giant larvaceans can contribute to the vertical flux of microplastics through the rapid sinking of fecal pellets and discarded houses. Larvaceans, and potentially other abundant pelagic filter feeders, may thus comprise a novel biological transport mechanism delivering microplastics from surface waters, through the water column, and to the seafloor. -

Effect of Appendicularians and Copepods on Bacterioplankton Composition and Growth in the English Channel
AQUATIC MICROBIAL ECOLOGY Vol. 32: 39–46, 2003 Published May 12 Aquat Microb Ecol Effect of appendicularians and copepods on bacterioplankton composition and growth in the English Channel Mikhail V. Zubkov1,*, Angel López-Urrutia2 1George Deacon Division for Ocean Processes, Southampton Oceanography Centre, University of Southampton, Empress Dock, Southampton SO14 3ZH, United Kingdom 2Plymouth Marine Laboratory, Prospect Place, Plymouth PL1 3DH, United Kingdom ABSTRACT: We compared the effects of the presence of the appendicularian Oikopleura dioica and the copepods Acartia clausii and Calanus helgolandicus on the coastal bacterioplankton community off Plymouth. Mesozooplankton were added to water samples and bacterioplankton growth was monitored by flow cytometry. Phylogenetic composition of bacterioplankton was analysed using flu- orescence in situ hybridisation (FISH) with rRNA-targeted oligonucleotide probes. The bacterio- plankton composition did not change in the presence of either appendicularians or copepods, and generally the same proportions of bacterioplankton groups were determined. In late spring, 15 ± 2% of cells hybridised with a probe specific to the Kingdom Archaea. The majority of cells (88 ± 2%) belonged to the Kingdom Bacteria, and 86% of cells were identified using group-specific probes. The Cytophage-Flavobacterium cluster dominated the community, comprising 64 ± 0.5% of cells. The γ- proteobacteria were the second abundant group, comprising 11 ± 0.5% of cells, and the SAR86 clus- ter of γ-proteobacteria accounted for 6 ± 5%. The α-proteobacteria comprised 10 ± 5% of bacterio- plankton, and the Roseobacteria related cluster represented 9 ± 3% of cells. The reduction of bacterioplankton growth caused by appendicularian bacterivory was 0.4 to 14% ind.–1 l–1, and the total appendicularian population could reduce bacterial growth in coastal waters in late spring- summer by up to 9%. -

National Monitoring Program for Biodiversity and Non-Indigenous Species in Egypt
National monitoring program for biodiversity and non-indigenous species in Egypt January 2016 1 TABLE OF CONTENTS page Acknowledgements 3 Preamble 4 Chapter 1: Introduction 8 Overview of Egypt Biodiversity 37 Chapter 2: Institutional and regulatory aspects 39 National Legislations 39 Regional and International conventions and agreements 46 Chapter 3: Scientific Aspects 48 Summary of Egyptian Marine Biodiversity Knowledge 48 The Current Situation in Egypt 56 Present state of Biodiversity knowledge 57 Chapter 4: Development of monitoring program 58 Introduction 58 Conclusions 103 Suggested Monitoring Program Suggested monitoring program for habitat mapping 104 Suggested marine MAMMALS monitoring program 109 Suggested Marine Turtles Monitoring Program 115 Suggested Monitoring Program for Seabirds 117 Suggested Non-Indigenous Species Monitoring Program 121 Chapter 5: Implementation / Operational Plan 128 Selected References 130 Annexes 141 2 AKNOWLEGEMENTS 3 Preamble The Ecosystem Approach (EcAp) is a strategy for the integrated management of land, water and living resources that promotes conservation and sustainable use in an equitable way, as stated by the Convention of Biological Diversity. This process aims to achieve the Good Environmental Status (GES) through the elaborated 11 Ecological Objectives and their respective common indicators. Since 2008, Contracting Parties to the Barcelona Convention have adopted the EcAp and agreed on a roadmap for its implementation. First phases of the EcAp process led to the accomplishment of 5 steps of the scheduled 7-steps process such as: 1) Definition of an Ecological Vision for the Mediterranean; 2) Setting common Mediterranean strategic goals; 3) Identification of an important ecosystem properties and assessment of ecological status and pressures; 4) Development of a set of ecological objectives corresponding to the Vision and strategic goals; and 5) Derivation of operational objectives with indicators and target levels. -

Title GENERAL CONSIDERATION on JAPANESE APPENDICULARIAN
GENERAL CONSIDERATION ON JAPANESE Title APPENDICULARIAN FAUNA Author(s) Tokioka, Takasi PUBLICATIONS OF THE SETO MARINE BIOLOGICAL Citation LABORATORY (1955), 4(2-3): 251-261 Issue Date 1955-05-30 URL http://hdl.handle.net/2433/174523 Right Type Departmental Bulletin Paper Textversion publisher Kyoto University GENERAL CONSIDERATION ON JAPANESE APPENDICULARIAN FAUNA') T AKASI TOKIOKA Seto Marine Biological Laboratory, Sirahama With 6 Text-figures I. Species Occlilrring ilil Japanese Waters and the Neighbolilring Seas. So far as I am aware, the following 34 species of appendicularians are found in reports dealing with this animal group occurring in Japanese waters and the neigh bouring seas. Oikopleuridae Oikopleura (Coecaria) longicauda VoGT (A, T,, T 2 , T 3 ) Oikopleura (Coecaria) fusiformis FoL (A, T,, T 2 , Ts) Oikopleura (Coecaria) intermedia LOHMANN ( =Oik. microstoma AIDA) (A, T,, T 2 ) *Oikopleura (Coecaria) cornutogastra AIDA (A, T 2 , T 3) Oikopleura (Coecaria) graciloides LOHMANN & BuCKMANN (T2 ) Oikopleura (Coecaria) gracilis LOHMANN (T3 ) Oikopleura ( Vexillaria) dioica FoL (A, T,, T 2 , T 3 ) Oikopleura (Vexillaria) rufescens FoL (A, T,, T 2 , T 3) Oikopleura ( Vexillaria) parva LoHMANN (T3 ) Oikopleura (Vexillaria) cophocerca GEGENBAUR (T1 , T 2 , T 3) Oikopleura ( Vexillaria) albicans (LEUCKART) (T,) Oikopleura ( Vexillaria) labradoriensis LoHMANN (T,, T 3 ) *Oikopleura ( Vexillaria) chamissonis MERTENS (MERTENS 1831) Megalocercus huxleyi (RITTER) (=Oik. megastoma AIDA) (A, T,, T 2 , T 3 ) Stegosoma magnum (LANGERHANS) (A, T 1 , T 2 , T 3 ) *Chunopleura microgaster LOHMANN (T,) Pelagopleura verticalis (LoHMANN) (T2 ) *Pelagopleura sp. (T,, T 3 ) Fritillaridae Fritillaria (Acrocercus) haplostoma FoL (A, T,, T 2 , Ts) ----------------------------------·------- 1) Contributions from the Seto Marine Biological Laboratory, No. -

Species Composition and Horizontal Distribution of the Appendicularian Community in Waters Adjacent to the Kuroshio in Winter–Early Spring
Plankton Benthos Res 3(3): 152–164, 2008 Plankton & Benthos Research © The Plankton Society of Japan Species composition and horizontal distribution of the appendicularian community in waters adjacent to the Kuroshio in winter–early spring KIYOTAKA HIDAKA* National Research Institute of Fisheries Science, Fisheries Research Agency, 2–12–4 Fukuura, Kanazawa, Yokohama, Kanagawa, 236–8648 Japan Received 10 December 2007; Accepted 10 April 2008 Abstract: Species composition and horizontal distribution of appendicularians were investigated in waters adjacent to the Kuroshio, south of Honshu in winter–early spring. Twenty-one species belonging to 5 genera were found and the species composition was characterized by a strong dominance of Oikopleura longicauda and occasional dense occur- rence of Fritillaria pellucida. Oikopleura longicauda was distributed rather uniformly throughout the study area while F. pellucida was usually only sparsely distributed, although it was sometimes found in dense concentrations at the sta- tions close to the Kuroshio axis. Occurrence of O. dioica, a well-studied neritic species, was limited to the stations with a relatively shallow bottom and its relative importance in terms of abundance was small. Generally, the appendicularian biomass was less than one tenth of the copepod biomass in carbon. Nevertheless, the role of appendicularians in terms of secondary production was estimated to be smaller but comparable in magnitude to that of copepods, 1.7–202.3 mgC mϪ2 dϪ1 in the former and 30.0–185.2 mgC mϪ2 dϪ1 in the latter, because of their higher somatic growth rates and house production. Clearance rate of appendicularians was also estimated to be comparable to copepods, 3.1–591.3 L mϪ2 hϪ1 in the former and 49.0–462.1 L mϪ2 hϪ1 in the latter, owing to their much higher weight specific clearance rates. -

Spatial Distribution Patterns of Appendicularians in the Drake Passage: Potential Indicators of Water Masses?
diversity Article Spatial Distribution Patterns of Appendicularians in the Drake Passage: Potential Indicators of Water Masses? Marcin Kalarus 1 and Anna Panasiuk 2,* 1 Department of Aquatic Ecology, Maritime Institute, Gdynia Maritime University, Smoluchowskiego 1/3, 80-214 Gda´nsk,Poland; [email protected] 2 Department of Marine Plankton Research, Faculty of Oceanography and Geography, Institute of Oceanography, University of Gda´nsk,Av. Marszałka J. Piłsudskiego 46, 81-378 Gdynia, Poland * Correspondence: [email protected]; Tel.: +485-8523-6844 Abstract: Appendicularians are one of the most common animals found within zooplankton as- semblages. They play a very important role as filter feeders but are, unfortunately, inconsistently reported in the Antarctic literature. The present paper attempts to describe the zonal diversity of appendicularians and the main environmental factors influencing their communities in the Drake Passage. Samples were collected during Antarctic summer in 2009–2010. A total of eight species of larvaceans were identified. Fritillaria borealis was the species found in the highest numbers in almost the entire studied area, and was observed at all sampling stations. The distributions of other taxa were limited to specific hydrological zones and hydrological conditions. F. fraudax and Oikopleura gaussica were typical of the areas between the Polar Front and the Subantarctic Front zones, and their distributions were significantly correlated with temperature and salinity, likely making them good indicator species. The F. fusiformis distribution was strictly related to South American waters. In summary, temperature was the strongest environmental factor influencing the larvacean community structure in the Drake Passage, and we also found that testing environmental factors on larvaceans Citation: Kalarus, M.; Panasiuk, A. -

A Chordate Species Lacking Nodal Utilizes Calcium Oscillation and Bmp for Left–Right Patterning
A chordate species lacking Nodal utilizes calcium oscillation and Bmp for left–right patterning Takeshi A. Onumaa,1, Momoko Hayashia, Fuki Gyojab, Kanae Kishia, Kai Wanga, and Hiroki Nishidaa aDepartment of Biological Sciences, Graduate School of Science, Osaka University, 560-0043 Toyonaka, Osaka, Japan; and bMarine Genomics Unit, Okinawa Institute of Science and Technology Graduate University, 904-0495 Onna, Okinawa, Japan Edited by Edward M. De Robertis, David Geffen School of Medicine, University of California, Los Angeles, CA, and approved December 20, 2019 (received for review September 27, 2019) Larvaceans are chordates with a tadpole-like morphology. In neurula by ciliary movement (27, 28). These are considered to contrast to most chordates of which early embryonic morphology is be biological cues that induce the left-sided expression of Nodal,a bilaterally symmetric and the left–right (L–R) axis is specified by the member of the TGF-β family, leading to the L–R asymmetry of Nodal pathway later on, invariant L–R asymmetry emerges in four- Deuterostomia (e.g., vertebrates) and Lophotrochozoa (e.g., pond cell embryos of larvaceans. The asymmetric cell arrangements exist snails). However, these mechanisms are not universal. For ex- through development of the tailbud. The tail thus twists 90° in a ample, motile cilia are not associated with L–R patterning in the counterclockwise direction relative to the trunk, and the tail nerve pond snail (29, 30), and Nodal is lost in Ecdysozoa (e.g., nem- cord localizes on the left side. Here, we demonstrate that larvacean atodes and flies). This diversity of symmetry-breaking mecha- embryos have nonconventional L–R asymmetries: 1) L- and R-cells of nisms suggests that there may be organisms with novel strategies 2+ the two-cell embryo had remarkably asymmetric cell fates; 2) Ca for L–R patterning.