Control of Tissue Development and Cell Diversity by Cell Cycle-Dependent Transcriptional Filtering Maria Abou Chakra, Ruth Isserlin, Thinh N Tran, Gary D Bader*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Appendicularia of CTAW
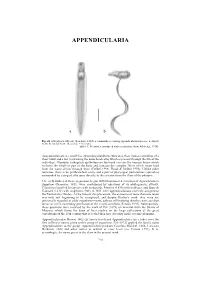
APPENDICULARIA APPENDICULARIA a b Fig. 26. Oikopleura albicans (Leuckart, 1854), a commonly occurring appendicularian species: a, dorsal view; b, lateral view. (Scale bar = 0.1 mm). [after T. Prentiss, reproduced with permission, from Alldredge 1976] Appendicularians are small free swimming planktonic tunicates, their bodies consisting of a short trunk and a tail (containing the notochord cells) which is present through the life of the individual. Glandular (oikoplast) epithelium on the trunk secretes the mucous house which encloses the whole or part of the body and contains the complex filters which strain food from the water driven through them (Deibel 1998; Flood & Deibel 1998). Unlike other tunicates, there is no peribranchial cavity and a pair of pharyngeal perforations (spiracles) surrounded by a ring of cilia open directly to the exterior from the floor of the pharynx. The early studies of these organisms, begun with Chamisso's description of Appendicularia flagellum Chamisso, 1821, were confounded by questions of its phylogenetic affinity. Chamisso classified his species with medusoids, Mertens (1830) with molluscs, and Quoy & Gaimard (1833) with zoophytes. Only in 1851 were appendicularians correctly assigned to the Tunicata by Huxley. At the time of this placement, the existence of more than one taxon was only just beginning to be recognised, and despite Huxley's work, they were not universally regarded as adult organisms—some authors still insisting that they were ascidian larvae or a free swimming generation of the sessile ascidians (Fenaux 1993). Subsequently, these questions were resolved by the work of Fol (1872) on material from the Straits of Messina, which forms the basis of later studies on the large collections of the great expeditions of the 19th century that revealed their true diversity in the oceanic plankton. -

Brains of Primitive Chordates 439
Brains of Primitive Chordates 439 Brains of Primitive Chordates J C Glover, University of Oslo, Oslo, Norway although providing a more direct link to the evolu- B Fritzsch, Creighton University, Omaha, NE, USA tionary clock, is nevertheless hampered by differing ã 2009 Elsevier Ltd. All rights reserved. rates of evolution, both among species and among genes, and a still largely deficient fossil record. Until recently, it was widely accepted, both on morpholog- ical and molecular grounds, that cephalochordates Introduction and craniates were sister taxons, with urochordates Craniates (which include the sister taxa vertebrata being more distant craniate relatives and with hemi- and hyperotreti, or hagfishes) represent the most chordates being more closely related to echinoderms complex organisms in the chordate phylum, particu- (Figure 1(a)). The molecular data only weakly sup- larly with respect to the organization and function of ported a coherent chordate taxon, however, indicat- the central nervous system. How brain complexity ing that apparent morphological similarities among has arisen during evolution is one of the most chordates are imposed on deep divisions among the fascinating questions facing modern science, and it extant deuterostome taxa. Recent analysis of a sub- speaks directly to the more philosophical question stantially larger number of genes has reversed the of what makes us human. Considerable interest has positions of cephalochordates and urochordates, pro- therefore been directed toward understanding the moting the latter to the most closely related craniate genetic and developmental underpinnings of nervous relatives (Figure 1(b)). system organization in our more ‘primitive’ chordate relatives, in the search for the origins of the vertebrate Comparative Appearance of Brains, brain in a common chordate ancestor. -

Global LC/MS Metabolomics Profiling of Calcium Stressed and Immunosuppressant Drug Treated Saccharomyces Cerevisiae
Metabolites 2013, 3, 1102-1117; doi:10.3390/metabo3041102 OPEN ACCESS metabolites ISSN 2218-1989 www.mdpi.com/journal/metabolites/ Article Global LC/MS Metabolomics Profiling of Calcium Stressed and Immunosuppressant Drug Treated Saccharomyces cerevisiae Stefan Jenkins 1,2,3,*, Steven M. Fischer 2, Lily Chen 3 and Theodore R. Sana 2 1 Life Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA 2 Agilent Technologies, Life Sciences, Diagnostics and Applied Markets, Santa Clara, CA 95051, USA; E-Mails: [email protected] (S.M.F.); [email protected] (T.R.S.) 3 Biology Department, San Francisco State University, San Francisco, CA 94132, USA; E-Mail: [email protected] * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-650-922-5046; Fax: +1-510-486-4545. Received: 15 October 2013; in revised form: 20 November 2013 / Accepted: 25 November 2013 / Published: 6 December 2013 Abstract: Previous studies have shown that calcium stressed Saccharomyces cerevisiae, challenged with immunosuppressant drugs FK506 and Cyclosporin A, responds with comprehensive gene expression changes and attenuation of the generalized calcium stress response. Here, we describe a global metabolomics workflow for investigating the utility of tracking corresponding phenotypic changes. This was achieved by efficiently analyzing relative abundance differences between intracellular metabolite pools from wild-type and calcium stressed cultures, with and without prior immunosuppressant drugs exposure. We used pathway database content from WikiPathways and YeastCyc to facilitate the projection of our metabolomics profiling results onto biological pathways. A key challenge was to increase the coverage of the detected metabolites. This was achieved by applying both reverse phase (RP) and aqueous normal phase (ANP) chromatographic separations, as well as electrospray ionization (ESI) and atmospheric pressure chemical ionization (APCI) sources for detection in both ion polarities. -

And Macro-Level Network Metrics Unveils Top Communicative Gene Modules in Psoriasis
G C A T T A C G G C A T genes Article Using Micro- and Macro-Level Network Metrics Unveils Top Communicative Gene Modules in Psoriasis 1, 2 3 Reyhaneh Naderi y, Homa Saadati Mollaei , Arne Elofsson 3, , and Saman Hosseini Ashtiani * y 1 Department of Artificial Intelligence and Robotics, Faculty of Computer Engineering, Iran University of Science and Technology, Tehran 1684613114, Iran; [email protected] 2 Department of Advanced Sciences and Technology, Islamic Azad University Tehran Medical Sciences, Tehran 1916893813, Iran; [email protected] 3 Department of Biochemistry and Biophysics and Science for Life Laboratory, Stockholm University, 106 91 Stockholm, Sweden; [email protected] * Correspondence: [email protected]; Tel.: +46-762623644 These authors contributed equally to this work and should be considered joint first authors. y Received: 9 July 2020; Accepted: 6 August 2020; Published: 10 August 2020 Abstract: (1) Background: Psoriasis is a multifactorial chronic inflammatory disorder of the skin, with significant morbidity, characterized by hyperproliferation of the epidermis. Even though psoriasis’ etiology is not fully understood, it is believed to be multifactorial, with numerous key components. (2) Methods: In order to cast light on the complex molecular interactions in psoriasis vulgaris at both protein–protein interactions and transcriptomics levels, we studied a set of microarray gene expression analyses consisting of 170 paired lesional and non-lesional samples. Afterwards, a network analysis was conducted on the protein–protein interaction network of differentially expressed genes based on micro- and macro-level network metrics at a systemic level standpoint. (3) Results: We found 17 top communicative genes, all of which were experimentally proven to be pivotal in psoriasis, which were identified in two modules, namely the cell cycle and immune system. -

Biological Pathways
Biological pathways Bing Zhang Department of Biomedical Informatics Vanderbilt University [email protected] Biological pathway A biological pathway is a series of actions among molecules in a cell that leads to a certain product or a change in a cell. Different types of biological pathways Signal transduction pathways Gene regulation pathways Metabolic pathways 2 Applied Bioinformatics, Spring 2011 Outline Pathway databases Pathway assembly and editing Pathway mapping and enrichment analysis 3 Applied Bioinformatics, Spring 2011 Selected pathway databases KEGG Kyoto Encyclopedia of Genes and Genomes http://www.genome.jp/kegg/pathway.html Reactome Ontario Institute for Cancer Research, Cold Spring Harbor Laboratory, New York University School of Medicine and The European Bioinformatics Institute http://www.reactome.org/ WikiPathways University of Maastricht and UCSF http://www.wikipathways.org/ 4 Applied Bioinformatics, Spring 2011 KEGG: data model Molecular building blocks KEGG GENES: genes and proteins generated by genome sequencing projects KEGG ORTHOLOGY: orthology (KO) groups KEGG COMPOUND: small molecules KEGG REACTIONS: biochemical reactions KEGG PATHWAY: pathway maps Created in a general way to be applicable to all organisms, in terms of the orthologs defined by KO groups Organism-specific pathways can be generated by converting KO groups to gene identifiers in a given organism 5 Applied Bioinformatics, Spring 2011 KEGG: TCA cycle 6 Applied Bioinformatics, Spring 2011 Reactome: data model Entities -

Confirmation of Pathogenic Mechanisms by SARS-Cov-2–Host
Messina et al. Cell Death and Disease (2021) 12:788 https://doi.org/10.1038/s41419-021-03881-8 Cell Death & Disease ARTICLE Open Access Looking for pathways related to COVID-19: confirmation of pathogenic mechanisms by SARS-CoV-2–host interactome Francesco Messina 1, Emanuela Giombini1, Chiara Montaldo1, Ashish Arunkumar Sharma2, Antonio Zoccoli3, Rafick-Pierre Sekaly2, Franco Locatelli4, Alimuddin Zumla5, Markus Maeurer6,7, Maria R. Capobianchi1, Francesco Nicola Lauria1 and Giuseppe Ippolito 1 Abstract In the last months, many studies have clearly described several mechanisms of SARS-CoV-2 infection at cell and tissue level, but the mechanisms of interaction between host and SARS-CoV-2, determining the grade of COVID-19 severity, are still unknown. We provide a network analysis on protein–protein interactions (PPI) between viral and host proteins to better identify host biological responses, induced by both whole proteome of SARS-CoV-2 and specific viral proteins. A host-virus interactome was inferred, applying an explorative algorithm (Random Walk with Restart, RWR) triggered by 28 proteins of SARS-CoV-2. The analysis of PPI allowed to estimate the distribution of SARS-CoV-2 proteins in the host cell. Interactome built around one single viral protein allowed to define a different response, underlining as ORF8 and ORF3a modulated cardiovascular diseases and pro-inflammatory pathways, respectively. Finally, the network-based approach highlighted a possible direct action of ORF3a and NS7b to enhancing Bradykinin Storm. This network-based representation of SARS-CoV-2 infection could be a framework for pathogenic evaluation of specific 1234567890():,; 1234567890():,; 1234567890():,; 1234567890():,; clinical outcomes. -

Spatial Distribution Patterns of Appendicularians in the Drake Passage: Potential Indicators of Water Masses?
diversity Article Spatial Distribution Patterns of Appendicularians in the Drake Passage: Potential Indicators of Water Masses? Marcin Kalarus 1 and Anna Panasiuk 2,* 1 Department of Aquatic Ecology, Maritime Institute, Gdynia Maritime University, Smoluchowskiego 1/3, 80-214 Gda´nsk,Poland; [email protected] 2 Department of Marine Plankton Research, Faculty of Oceanography and Geography, Institute of Oceanography, University of Gda´nsk,Av. Marszałka J. Piłsudskiego 46, 81-378 Gdynia, Poland * Correspondence: [email protected]; Tel.: +485-8523-6844 Abstract: Appendicularians are one of the most common animals found within zooplankton as- semblages. They play a very important role as filter feeders but are, unfortunately, inconsistently reported in the Antarctic literature. The present paper attempts to describe the zonal diversity of appendicularians and the main environmental factors influencing their communities in the Drake Passage. Samples were collected during Antarctic summer in 2009–2010. A total of eight species of larvaceans were identified. Fritillaria borealis was the species found in the highest numbers in almost the entire studied area, and was observed at all sampling stations. The distributions of other taxa were limited to specific hydrological zones and hydrological conditions. F. fraudax and Oikopleura gaussica were typical of the areas between the Polar Front and the Subantarctic Front zones, and their distributions were significantly correlated with temperature and salinity, likely making them good indicator species. The F. fusiformis distribution was strictly related to South American waters. In summary, temperature was the strongest environmental factor influencing the larvacean community structure in the Drake Passage, and we also found that testing environmental factors on larvaceans Citation: Kalarus, M.; Panasiuk, A. -

A Chordate Species Lacking Nodal Utilizes Calcium Oscillation and Bmp for Left–Right Patterning
A chordate species lacking Nodal utilizes calcium oscillation and Bmp for left–right patterning Takeshi A. Onumaa,1, Momoko Hayashia, Fuki Gyojab, Kanae Kishia, Kai Wanga, and Hiroki Nishidaa aDepartment of Biological Sciences, Graduate School of Science, Osaka University, 560-0043 Toyonaka, Osaka, Japan; and bMarine Genomics Unit, Okinawa Institute of Science and Technology Graduate University, 904-0495 Onna, Okinawa, Japan Edited by Edward M. De Robertis, David Geffen School of Medicine, University of California, Los Angeles, CA, and approved December 20, 2019 (received for review September 27, 2019) Larvaceans are chordates with a tadpole-like morphology. In neurula by ciliary movement (27, 28). These are considered to contrast to most chordates of which early embryonic morphology is be biological cues that induce the left-sided expression of Nodal,a bilaterally symmetric and the left–right (L–R) axis is specified by the member of the TGF-β family, leading to the L–R asymmetry of Nodal pathway later on, invariant L–R asymmetry emerges in four- Deuterostomia (e.g., vertebrates) and Lophotrochozoa (e.g., pond cell embryos of larvaceans. The asymmetric cell arrangements exist snails). However, these mechanisms are not universal. For ex- through development of the tailbud. The tail thus twists 90° in a ample, motile cilia are not associated with L–R patterning in the counterclockwise direction relative to the trunk, and the tail nerve pond snail (29, 30), and Nodal is lost in Ecdysozoa (e.g., nem- cord localizes on the left side. Here, we demonstrate that larvacean atodes and flies). This diversity of symmetry-breaking mecha- embryos have nonconventional L–R asymmetries: 1) L- and R-cells of nisms suggests that there may be organisms with novel strategies 2+ the two-cell embryo had remarkably asymmetric cell fates; 2) Ca for L–R patterning. -

Ascidian News #82 December 2018
ASCIDIAN NEWS* Gretchen Lambert 12001 11th Ave. NW, Seattle, WA 98177 206-365-3734 [email protected] home page: http://depts.washington.edu/ascidian/ Number 82 December 2018 A big thank-you to all who sent in contributions. There are 85 New Publications listed at the end of this issue. Please continue to send me articles, and your new papers, to be included in the June 2019 issue of AN. It’s never too soon to plan ahead. *Ascidian News is not part of the scientific literature and should not be cited as such. NEWS AND VIEWS 1. From Stefano Tiozzo ([email protected]) and Remi Dumollard ([email protected]): The 10th Intl. Tunicata Meeting will be held at the citadel of Saint Helme in Villefranche sur Mer (France), 8- 12 July 2019. The web site with all the information will be soon available, save the date! We are looking forward to seeing you here in the Riviera. A bientôt! Remi and Stefano 2. The 10th Intl. Conference on Marine Bioinvasions was held in Puerto Madryn, Patagonia, Argentina, October 16-18. At the conference website (http://www.marinebioinvasions.info/index) the program and abstracts in pdf can be downloaded. Dr. Rosana Rocha presented one of the keynote talks: "Ascidians in the anthropocene - invasions waiting to happen". See below under Meetings Abstracts for all the ascidian abstracts; my thanks to Evangelina Schwindt for compiling them. The next (11th) meeting will be in Annapolis, Maryland, organized by Greg Ruiz, Smithsonian Invasions lab, date to be determined. 3. Conference proceedings of the May 2018 Invasive Sea Squirt Conference will be peer reviewed and published in a special issue of the REABIC journal Management of Biological Invasions. -

Post-Graduate Diploma Program in Personalized Genomic Medicine” for Skill Development (DBT Sponsored Scheme)
UNIVERSITY OF MYSORE Syllabus for “Post-graduate Diploma Program in Personalized Genomic Medicine” For Skill Development (DBT Sponsored Scheme) FCBCS-CAGP SYSTEM Department of Studies in Genetics and Genomics University of Mysore Manasagangotri, Mysuru – 570 006 2019-20 1 PREAMBLE: The Information and potential use of genomic discoveries are no longer issues left for scientists and medical professionals to handle, but have become ones for the public at large. Rarely a day passes without genomics related story reported in the media. The proposed diploma program is designed to provide advanced knowledge dissemination in the field of genome sciences, applications and laboratory skills needed for molecular diagnostics and precision medicine procedures conducted in a clinical or research environment. The program is intended for those individuals who wish to enhance their laboratory expertise and knowledge in molecular-based methods. Precision medicine has the potential to fundamentally change how health care is practiced, but requires a trained health care workforce that understands the complexities of this field. One important component of precision medicine is the use of an individual’s genomic information to offer targeted treatment, tailored to the individual. Our course aims to provide participants with advanced knowledge of genomics, an overview of the clinical applications of genomic medicine, the skills to evaluate the clinical validity and utility of new tests, and an appreciation of the associated ethical and social issues inherent in this field. The course is geared towards individuals with a background in the biological sciences and a basic understanding of genetics. It is designed to be succinct and clinically focused, offering both conceptual and practical information about real-world applications of genomics. -

Oikopleura Dioica
To: [email protected] o ABSTRACT We propose that a BAC library should be constructed for the larvacean urochordate Oikopleura dioica. This animal may be the most similar among extant animals to the last common ancestor of all chordates, and thus, its investigation can illuminate the mechanisms causing the vertebrate genome expansion, the principles that guide the evolution of functions of duplicate genes in development, and the origin of vertebrate developmental innovations. Oikopleura dioica has a genome of about 70 Mb, the smallest of any chordate. o IMPORTANCE OF THE ORGANISM TO BIOMEDICAL OR BIOLOGICAL RESEARCH. The Animal. Oikopleura dioica is a 3 mm long urochordate, an abundant filter-feeder in marine plankton. Oikopleura and other members of its phylogenetic class, the larvaceans (or appendicularians) remain free swimming and tadpole-like into sexual maturity. Because larvaceans are basally diverging among the urochordates, this free-swimming life style may have been ancestral (Wada 1998). Larvaceans have more functional organ systems than do ascidian larvae; their embryos, tadpole larvae, and adults are transparent, which facilitates observations on all aspects of development (Fig. 1); embryonic development takes only 14 hours; and the generation time is less than two weeks. Furthermore, Oikopleura dioica is readily available from plankton tows for much of the year off Northern hemisphere coasts. Oikopleura dioica can be among the most abundant animals in offshore waters, even more than copepods, amounting to several animals per liter (Dagg & Ortner 1994; Capitano & Esnal 1998). With Oikopleura, one can perform single-pair matings, and culture the offspring in the lab continuously in the presence of fresh sea water (Fenaux & Gorsky, 1983; Bassham, unpubl). -

Signaling in the Innate Immune Response
Signaling in the innate immune response The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Kim, Dennis H. “Signaling in the Innate Immune Response.” WormBook (December 2015): 1–51 © 2015 WormBook As Published http://dx.doi.org/10.1895/wormbook.1.83.2 Publisher WormBook Version Final published version Citable link http://hdl.handle.net/1721.1/109292 Terms of Use Creative Commons Attribution Detailed Terms http://creativecommons.org/licenses/by/2.5/ Signaling in the innate immune response Dennis H. Kim1§ and Jonathan J. Ewbank2§ 1Department of Biology, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA 2 Centre d’Immunologie de Marseille-Luminy, Aix Marseille Université UM2, INSERM, U1104, CNRS UMR7280, 13288 Marseille, France. Edited by Iva Greenwald Last revised September 16, 2015 §To whom correspondence should be addressed. E-mail: [email protected] or [email protected] WormBook Early Online, published on December 22, 2015 as doi: 10.1895/wormbook.1.83.2. 1 1. Introduction The maintenance of cellular and organismal homeostasis in the face of changes in the environment is essential for survival. Pathogenic microorganisms represent one type of environmental challenge. Plants and animals have developed immune systems to counter the threat of infection. The innate immune system is an ancient system of host defense against microbial pathogens present across a very broad range of species. Genetic studies of innate immunity in Drosophila and mammals have revealed an evolutionary conservation of signaling mechanisms (Medzhitov and Janeway, 1998), which has been corroborated in part by the study of innate immunity in Caenorhabditis elegans.