Putting SMYD3 on The
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Epigenetic Loss of the RNA Decapping Enzyme NUDT16 Mediates C-MYC Activation in T-Cell Acute Lymphoblastic Leukemia
OPEN Leukemia (2017) 31, 1622–1657 www.nature.com/leu LETTERS TO THE EDITOR Epigenetic loss of the RNA decapping enzyme NUDT16 mediates C-MYC activation in T-cell acute lymphoblastic leukemia Leukemia (2017) 31, 1622–1625; doi:10.1038/leu.2017.99 Having found the aforementioned NUDT16 CpG island methy- lation profiles, we studied in greater detail their association with the possible transcriptional inactivation of the NUDT16 gene at the RNA and protein levels in leukemia cell lines. We first It is possible that the occurrence of intrinsic defects in RNA performed bisulfite genomic sequencing of mutiple clones in the – processing pathways, such as RNA decapping,1 3 contribute to the T-cell Acute Lymphoblastic Leukemia (T-ALL) cell lines CCRF-CEM, distorted RNA landscapes of cancer cells. After transcription by Jurkat, MOLT-4 and MOLT-16 using primers that encompassed the RNA polymerase II, RNA molecules are equipped with a 5´-end transcription start site-associated CpG island and confirmed the N7-methyl guanosine (m7G)-cap. This m7G-cap is essential for hypermethylated status of the 5′-end region of NUDT16 in translation, stabilizing the RNA molecule and protecting it from comparison to normal T lymphocytes (Figure 1b), validating the – exonucleolytic breakdown.1 3 For RNA decay to occur the DNA methylation patterns obtained by the microarray approach m7G-cap first needs to be removed. This process is known as (Supplementary Figure S2). In contrast, normal T lymphocytes, the – decapping.1 3 The decapping mRNA 2 (DCP2) enzyme,4 also T-ALL cell lines KOPN-8, REH and RS4;11 and the leukemia cell known as the nucleoside diphosphate-linked moiety X motif lines HL-60 and K562 derived from myeloid lineage were all found 20 (NUDT20), was originally thought to be the only mammalian to be unmethylated (Figure 1b; Supplementary Figure S2). -

STAT3 Promotes Chronic Lymphocytic Leukemia Progression Through Upregulating SMYD3 Expression
Basic research Haematology STAT3 promotes chronic lymphocytic leukemia progression through upregulating SMYD3 expression Wei Ma1, Yingying Zhang2, Yu Qi3, Shidong Guo4 1Hematology and Oncology Department, Dongzhimen Hospital, Beijing University Corresponding author: of Chinese Medicine, Beijing, China Shidong Guo 2Scientific Research Department, Dongzhimen Hospital, Beijing University of Chinese Emergency Department Medicine, Beijing, China China-Japan 3Nursing Department, Dongzhimen Hospital, Beijing University of Chinese Medicine, Friendship Hospital Beijing, China No. 2 Sakura East St 4Emergency Department, China-Japan Friendship Hospital, Beijing, China Chaoyang District Beijing 100029, China Submitted: 3 May 2018 Phone/fax: Accepted: 14 July 2018 +86 010 84205185 E-mail: [email protected] Arch Med Sci 2019; 15 (5): 1163–1175 DOI: https://doi.org/10.5114/aoms.2018.77733 Copyright © 2018 Termedia &Banach Abstract Introduction: This study was designed to investigate the roles of STAT3 and SMYD3 in chronic lymphocytic leukemia and the regulatory relationship be- tween STAT3 and SMYD3 in chronic lymphocytic leukemia. Material and methods: The expression of STAT3 and SMYD3 was deter- mined by RT-qPCR and western blot in chronic lymphocytic leukemia sam- ples and cells (MEC1, CLL). Small interfering RNA was used to knock down the mRNA level of STAT3 and the pcDNA3.1-SMYD3 plasmid was used to construct a SMYD3 overexpression model. An MTT assay was performed to evaluate cell proliferation. A transwell assay was used to detect cell invasion ability. Afterwards, a luciferase reporter assay and chromatin immunopre- cipitation experiment (ChIP assay) were applied to confirm the correlation between STAT3 and SMYD3. Results: STAT3 was highly expressed in chronic lymphocytic leukemia mono- nuclear cells and cancerous cell lines. -
C a B D G E F
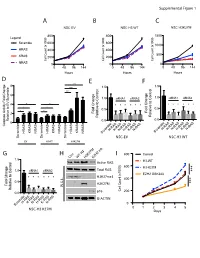
Supplemental Figure 1 A B C NSC EV NSC H3 WT NSC H3K27M 800 800 1500 ) ) Legend ) Scramble 600 600 x1000 x1000 x1000 ( ( ( 1000 t t t n n HRAS n 400 400 u u u o o o C C C 500 l l KRAS l 200 200 el el el C C C NRAS 0 0 0 0 48 96 144 0 48 96 144 0 48 96 144 Hours Hours Hours D *** E F 8 1.5 1.5 e *** l l *** ro ro t 6 t 1.0 siRNA1 siRNA2 1.0 siRNA1 siRNA2 Con Con o o t 4 * * t * * * * * * e * * * * * * e v i * * v i t 0.5 * * t 0.5 Fold Change 2 Fold Change Rela Rela Relative to EV Scrambl 0.0 0.0 0 Caspase Activity Fold Change H-RASK-RASN-RASH-RASK-RASN-RAS H-RASK-RASN-RASH-RASK-RASN-RAS KRAS KRAS KRAS HRAS NRAS HRAS NRAS HRAS NRAS Scramble Scramble Scramble Scramble Scramble NSC-EV NSC-H3 WT EV H3WT H3K27M G H I 800 Control Con WT H3 H3K27MEZH2 inb 1.5 l H3-WT Active RAS ro ) t 600 H3-K27M 1.0 siRNA1 siRNA2 Total RAS *** Con x1000 ( o EZH2 GSK343 t t * * * * * * H3K27me3 n e 400 u v i o t 0.5 *** H3K27M C Fold Change l W.C.L. el Rela p16 C 200 0.0 B-ACTIN H-RASK-RASN-RASH-RASK-RASN-RAS 0 Scramble NSC-H3 K27M 0 1 2 3 4 5 Days E C A 100 Cell Viability 100 25 50 75 Cell Viability 25 50 75 0 0 Fold Change MOCK MOCK Relative to Control Scramble * 0.0 0.2 0.4 0.6 0.8 1.0 1.2 MYC Scramble MYC MYC * PDGFRA Scramble siRNA PDGFRA MYC PDGFRA AURKA AURKA DIPG007siRNA2 PDGFRA NSC H3K27MsiRNA1and AURKA LAMTOR3 AURKA LAMTOR3 LAMTOR3 LIN28A LAMTOR3 LIN28A LIN28B LIN28A NSC-EV LIN28A MAP2K3 LIN28B LIN28B LIN28B MAP2K5 siRNA1 MAP2K3 MAP2K3 MAP3K2 MAP2K3 MAP3K7 MAP2K5 MAP2K5 MAPK7 MAP2K5 2 MAP3K2 siRNA1 siRNA2 MAP3K2 ZAK MAP3K7 MAP3K2 MAP3K7 MAPK7 MAP3K7 -

Datasheet PB0939 Anti-SMYD3 Antibody
Product datasheet Anti-SMYD3 Antibody Catalog Number: PB0939 BOSTER BIOLOGICAL TECHNOLOGY Special NO.1, International Enterprise Center, 2nd Guanshan Road, Wuhan, China Web: www.boster.com.cn Phone: +86 27 67845390 Fax: +86 27 67845390 Email: [email protected] Basic Information Product Name Anti-SMYD3 Antibody Gene Name SMYD3 Source Rabbit IgG Species Reactivity human,mouse,rat Tested Application WB,IHC-P Contents 500ug/ml antibody with PBS ,0.02% NaN3 , 1mg BSA and 50% glycerol. Immunogen A synthetic peptide corresponding to a sequence at the C-terminus of human SMYD3 (388-428aa QAMKNLRLAFDIMRVTHGREHSLIEDLILLLEECDANIRAS), different from the related mouse sequence by one amino acid. Purification Immunogen affinity purified. Observed MW 55KD Dilution Ratios Western blot: 1:500-2000 Immunohistochemistry(Paraffin-embedded Section): 1:50-400 (Boiling the paraffin sections in 10mM citrate buffer,pH6.0,or PH8.0 EDTA repair liquid for 20 mins is required for the staining of formalin/paraffin sections.) Optimal working dilutions must be determined by end user. Storage 12 months from date of receipt,-20℃ as supplied.6 months 2 to 8℃ after reconstitution. Avoid repeated freezing and thawing Background Information SET and MYND domain-containing protein 3 is a protein that in humans is encoded by the SMYD3 gene. The International Radiation Hybrid Mapping Consortium mapped the SMYD3 gene to chromosome 1. This gene encodes a histone methyltransferase which functions in RNA polymerase II complexes by an interaction with a specific RNA helicase. Multiple transcript variants encoding different isoforms have been found for this gene. Reference Anti-SMYD3 Antibody被引用在0文献中。 暂无引用 FOR RESEARCH USE ONLY. -

Characterization of the Small Molecule Kinase Inhibitor SU11248 (Sunitinib/ SUTENT in Vitro and in Vivo
TECHNISCHE UNIVERSITÄT MÜNCHEN Lehrstuhl für Genetik Characterization of the Small Molecule Kinase Inhibitor SU11248 (Sunitinib/ SUTENT in vitro and in vivo - Towards Response Prediction in Cancer Therapy with Kinase Inhibitors Michaela Bairlein Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ. -Prof. Dr. K. Schneitz Prüfer der Dissertation: 1. Univ.-Prof. Dr. A. Gierl 2. Hon.-Prof. Dr. h.c. A. Ullrich (Eberhard-Karls-Universität Tübingen) 3. Univ.-Prof. A. Schnieke, Ph.D. Die Dissertation wurde am 07.01.2010 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 19.04.2010 angenommen. FOR MY PARENTS 1 Contents 2 Summary ................................................................................................................................................................... 5 3 Zusammenfassung .................................................................................................................................................... 6 4 Introduction .............................................................................................................................................................. 8 4.1 Cancer .............................................................................................................................................................. -

The Methyltransferase SMYD3 Mediates the Recruitment of Transcriptional Cofactors at the Myostatin and C-Met Genes and Regulates Skeletal Muscle Atrophy
Downloaded from genesdev.cshlp.org on October 2, 2021 - Published by Cold Spring Harbor Laboratory Press The methyltransferase SMYD3 mediates the recruitment of transcriptional cofactors at the myostatin and c-Met genes and regulates skeletal muscle atrophy Valentina Proserpio,1,3 Raffaella Fittipaldi,1,3 James G. Ryall,2 Vittorio Sartorelli,2,4 and Giuseppina Caretti1,4,5 1Department of Biosciences, University of Milan, 20133 Milan, Italy; 2Laboratory of Muscle Stem Cells and Gene Regulation, National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases, Bethesda, Maryland 20892, USA Elucidating the epigenetic mechanisms underlying muscle mass determination and skeletal muscle wasting holds the potential of identifying molecular pathways that constitute possible drug targets. Here, we report that the methyltransferase SMYD3 modulates myostatin and c-Met transcription in primary skeletal muscle cells and C2C12 myogenic cells. SMYD3 targets the myostatin and c-Met genes and participates in the recruitment of the bromodomain protein BRD4 to their regulatory regions through protein–protein interaction. By recruiting BRD4, SMYD3 favors chromatin engagement of the pause–release factor p-TEFb (positive transcription elongation factor) and elongation of Ser2-phosphorylated RNA polymerase II (PolIISer2P). Reducing SMYD3 decreases myostatin and c-Met transcription, thus protecting from glucocorticoid-induced myotube atrophy. Supporting functional relevance of the SMYD3/BRD4 interaction, BRD4 pharmacological -

A Cytoplasmic COMPASS Is Necessary for Cell Survival and Triple-Negative Breast Cancer Pathogenesis by Regulating Metabolism
Downloaded from genesdev.cshlp.org on September 27, 2021 - Published by Cold Spring Harbor Laboratory Press A cytoplasmic COMPASS is necessary for cell survival and triple-negative breast cancer pathogenesis by regulating metabolism Lu Wang,1 Clayton K. Collings,1 Zibo Zhao,1 Kira Alia Cozzolino,1,2 Quanhong Ma,3 Kaiwei Liang,1 Stacy A. Marshall,1 Christie C. Sze,1 Rintaro Hashizume,2 Jeffrey Nicholas Savas,2 and Ali Shilatifard1,4 1Department of Biochemistry and Molecular Genetics, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, USA; 2Department of Neurology, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, USA; 3Department of Neurosurgery, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, USA; 4Robert H. Lurie National Cancer Institute Comprehensive Cancer Center, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, USA Mutations and translocations within the COMPASS (complex of proteins associated with Set1) family of histone lysine methyltransferases are associated with a large number of human diseases, including cancer. Here we report that SET1B/COMPASS, which is essential for cell survival, surprisingly has a cytoplasmic variant. SET1B, but not its SET domain, is critical for maintaining cell viability, indicating a novel catalytic-independent role of SET1B/ COMPASS. Loss of SET1B or its unique cytoplasmic-interacting protein, BOD1, leads to up-regulation of expression of numerous genes modulating fatty acid metabolism, including ADIPOR1 (adiponectin receptor 1), COX7C, SDC4, and COQ7. Our detailed molecular studies identify ADIPOR1 signaling, which is inactivated in both obesity and human cancers, as a key target of SET1B/COMPASS. Collectively, our study reveals a cytoplasmic function for a member of the COMPASS family, which could be harnessed for therapeutic regulation of signaling in human dis- eases, including cancer. -

A Genome-Wide Screen in Mice to Identify Cell-Extrinsic Regulators of Pulmonary Metastatic Colonisation
FEATURED ARTICLE MUTANT SCREEN REPORT A Genome-Wide Screen in Mice To Identify Cell-Extrinsic Regulators of Pulmonary Metastatic Colonisation Louise van der Weyden,1 Agnieszka Swiatkowska, Vivek Iyer, Anneliese O. Speak, and David J. Adams Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SA, United Kingdom ORCID IDs: 0000-0002-0645-1879 (L.v.d.W.); 0000-0003-4890-4685 (A.O.S.); 0000-0001-9490-0306 (D.J.A.) ABSTRACT Metastatic colonization, whereby a disseminated tumor cell is able to survive and proliferate at a KEYWORDS secondary site, involves both tumor cell-intrinsic and -extrinsic factors. To identify tumor cell-extrinsic metastasis (microenvironmental) factors that regulate the ability of metastatic tumor cells to effectively colonize a metastatic tissue, we performed a genome-wide screen utilizing the experimental metastasis assay on mutant mice. colonisation Mutant and wildtype (control) mice were tail vein-dosed with murine metastatic melanoma B16-F10 cells and microenvironment 10 days later the number of pulmonary metastatic colonies were counted. Of the 1,300 genes/genetic B16-F10 locations (1,344 alleles) assessed in the screen 34 genes were determined to significantly regulate pulmonary lung metastatic colonization (15 increased and 19 decreased; P , 0.005 and genotype effect ,-55 or .+55). mutant While several of these genes have known roles in immune system regulation (Bach2, Cyba, Cybb, Cybc1, Id2, mouse Igh-6, Irf1, Irf7, Ncf1, Ncf2, Ncf4 and Pik3cg) most are involved in a disparate range of biological processes, ranging from ubiquitination (Herc1) to diphthamide synthesis (Dph6) to Rho GTPase-activation (Arhgap30 and Fgd4), with no previous reports of a role in the regulation of metastasis. -

The Lysine Methylase SMYD3 Modulates Mesendodermal Commitment During Development
cells Article The Lysine Methylase SMYD3 Modulates Mesendodermal Commitment during Development Raffaella Fittipaldi 1,†, Pamela Floris 1,† , Valentina Proserpio 1,2 , Franco Cotelli 1, Monica Beltrame 1 and Giuseppina Caretti 1,* 1 Department of Biosciences, University of Milan, Via Celoria 26, 20133 Milan, Italy; raffaella.fi[email protected] (R.F.); pamela.fl[email protected] (P.F.); [email protected] (V.P.); [email protected] (F.C.); [email protected] (M.B.) 2 Candiolo Cancer Institute, FPO-IRCCS, 10060 Candiolo, Italy * Correspondence: [email protected]; Tel.: +39-025-031-5002 † These authors contributed equally. Abstract: SMYD3 (SET and MYND domain containing protein 3) is a methylase over-expressed in cancer cells and involved in oncogenesis. While several studies uncovered key functions for SMYD3 in cancer models, the SMYD3 role in physiological conditions has not been fully elucidated yet. Here, we dissect the role of SMYD3 at early stages of development, employing mouse embryonic stem cells (ESCs) and zebrafish as model systems. We report that SMYD3 depletion promotes the induction of the mesodermal pattern during in vitro differentiation of ESCs and is linked to an upregulation of cardiovascular lineage markers at later stages. In vivo, smyd3 knockdown in zebrafish favors the upregulation of mesendodermal markers during zebrafish gastrulation. Overall, our study reveals that SMYD3 modulates levels of mesendodermal markers, both in development and in embryonic stem cell differentiation. Citation: Fittipaldi, R.; Floris, P.; Proserpio, V.; Cotelli, F.; Beltrame, M.; Keywords: embryonic stem cells; SMYD3; zebrafish; development Caretti, G. The Lysine Methylase SMYD3 Modulates Mesendodermal Commitment during Development. Cells 2021, 10, 1233. -

Molecular Signatures of Membrane Protein Complexes Underlying Muscular Dystrophy*□S
crossmark Research Author’s Choice © 2016 by The American Society for Biochemistry and Molecular Biology, Inc. This paper is available on line at http://www.mcponline.org Molecular Signatures of Membrane Protein Complexes Underlying Muscular Dystrophy*□S Rolf Turk‡§¶ʈ**, Jordy J. Hsiao¶, Melinda M. Smits¶, Brandon H. Ng¶, Tyler C. Pospisil‡§¶ʈ**, Kayla S. Jones‡§¶ʈ**, Kevin P. Campbell‡§¶ʈ**, and Michael E. Wright¶‡‡ Mutations in genes encoding components of the sar- The muscular dystrophies are hereditary diseases charac- colemmal dystrophin-glycoprotein complex (DGC) are re- terized primarily by the progressive degeneration and weak- sponsible for a large number of muscular dystrophies. As ness of skeletal muscle. Most are caused by deficiencies in such, molecular dissection of the DGC is expected to both proteins associated with the cell membrane (i.e. the sarco- reveal pathological mechanisms, and provides a biologi- lemma in skeletal muscle), and typical features include insta- cal framework for validating new DGC components. Es- bility of the sarcolemma and consequent death of the myofi- tablishment of the molecular composition of plasma- ber (1). membrane protein complexes has been hampered by a One class of muscular dystrophies is caused by mutations lack of suitable biochemical approaches. Here we present in genes that encode components of the sarcolemmal dys- an analytical workflow based upon the principles of pro- tein correlation profiling that has enabled us to model the trophin-glycoprotein complex (DGC). In differentiated skeletal molecular composition of the DGC in mouse skeletal mus- muscle, this structure links the extracellular matrix to the cle. We also report our analysis of protein complexes in intracellular cytoskeleton. -

Activation of Diverse Signalling Pathways by Oncogenic PIK3CA Mutations
ARTICLE Received 14 Feb 2014 | Accepted 12 Aug 2014 | Published 23 Sep 2014 DOI: 10.1038/ncomms5961 Activation of diverse signalling pathways by oncogenic PIK3CA mutations Xinyan Wu1, Santosh Renuse2,3, Nandini A. Sahasrabuddhe2,4, Muhammad Saddiq Zahari1, Raghothama Chaerkady1, Min-Sik Kim1, Raja S. Nirujogi2, Morassa Mohseni1, Praveen Kumar2,4, Rajesh Raju2, Jun Zhong1, Jian Yang5, Johnathan Neiswinger6, Jun-Seop Jeong6, Robert Newman6, Maureen A. Powers7, Babu Lal Somani2, Edward Gabrielson8, Saraswati Sukumar9, Vered Stearns9, Jiang Qian10, Heng Zhu6, Bert Vogelstein5, Ben Ho Park9 & Akhilesh Pandey1,8,9 The PIK3CA gene is frequently mutated in human cancers. Here we carry out a SILAC-based quantitative phosphoproteomic analysis using isogenic knockin cell lines containing ‘driver’ oncogenic mutations of PIK3CA to dissect the signalling mechanisms responsible for oncogenic phenotypes induced by mutant PIK3CA. From 8,075 unique phosphopeptides identified, we observe that aberrant activation of PI3K pathway leads to increased phosphorylation of a surprisingly wide variety of kinases and downstream signalling networks. Here, by integrating phosphoproteomic data with human protein microarray-based AKT1 kinase assays, we discover and validate six novel AKT1 substrates, including cortactin. Through mutagenesis studies, we demonstrate that phosphorylation of cortactin by AKT1 is important for mutant PI3K-enhanced cell migration and invasion. Our study describes a quantitative and global approach for identifying mutation-specific signalling events and for discovering novel signalling molecules as readouts of pathway activation or potential therapeutic targets. 1 McKusick-Nathans Institute of Genetic Medicine and Department of Biological Chemistry, Johns Hopkins University School of Medicine, 733 North Broadway, BRB 527, Baltimore, Maryland 21205, USA.