Characterization of Novel Genic SSR Markers in Linum Usitatissimum (L.) and Their Transferability Across Eleven Linum Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

FLORA from FĂRĂGĂU AREA (MUREŞ COUNTY) AS POTENTIAL SOURCE of MEDICINAL PLANTS Silvia OROIAN1*, Mihaela SĂMĂRGHIŢAN2
ISSN: 2601 – 6141, ISSN-L: 2601 – 6141 Acta Biologica Marisiensis 2018, 1(1): 60-70 ORIGINAL PAPER FLORA FROM FĂRĂGĂU AREA (MUREŞ COUNTY) AS POTENTIAL SOURCE OF MEDICINAL PLANTS Silvia OROIAN1*, Mihaela SĂMĂRGHIŢAN2 1Department of Pharmaceutical Botany, University of Medicine and Pharmacy of Tîrgu Mureş, Romania 2Mureş County Museum, Department of Natural Sciences, Tîrgu Mureş, Romania *Correspondence: Silvia OROIAN [email protected] Received: 2 July 2018; Accepted: 9 July 2018; Published: 15 July 2018 Abstract The aim of this study was to identify a potential source of medicinal plant from Transylvanian Plain. Also, the paper provides information about the hayfields floral richness, a great scientific value for Romania and Europe. The study of the flora was carried out in several stages: 2005-2008, 2013, 2017-2018. In the studied area, 397 taxa were identified, distributed in 82 families with therapeutic potential, represented by 164 medical taxa, 37 of them being in the European Pharmacopoeia 8.5. The study reveals that most plants contain: volatile oils (13.41%), tannins (12.19%), flavonoids (9.75%), mucilages (8.53%) etc. This plants can be used in the treatment of various human disorders: disorders of the digestive system, respiratory system, skin disorders, muscular and skeletal systems, genitourinary system, in gynaecological disorders, cardiovascular, and central nervous sistem disorders. In the study plants protected by law at European and national level were identified: Echium maculatum, Cephalaria radiata, Crambe tataria, Narcissus poeticus ssp. radiiflorus, Salvia nutans, Iris aphylla, Orchis morio, Orchis tridentata, Adonis vernalis, Dictamnus albus, Hammarbya paludosa etc. Keywords: Fărăgău, medicinal plants, human disease, Mureş County 1. -

Ethnobotanical Study of Medicinal Plants of Namal Valley, Salt Range, Pakistan - 4725
Shah et al.: Ethnobotanical study of medicinal plants of Namal Valley, Salt Range, Pakistan - 4725 - ETHNOBOTANICAL STUDY OF MEDICINAL PLANTS OF NAMAL VALLEY, SALT RANGE, PAKISTAN SHAH, A.1* – POUDEL, R. C.2 – ISHTIAQ, M.3 – SARVAT, R.1 – SHAHZAD, H.1 – ABBAS, A.1 – SHOAIB, S.1 – NUZHAT, R.1 – NOOR, U. D.1 – MAHMOODA, H.1 – SUMMAYA, A.1 – IFRA, A.1 – IHSAN, U.1 1Department of Botany, University of Sargodha, Sargodha-40100, Pakistan 2Nepal Academy of Science and Technology, Pātan-44700, Nepal 3Department of Botany, (Bhimber Campus), Mirpur University of Science & Technology Mirpur-10250 (AJK), Pakistan Corresponding author٭ e-mail: [email protected] ; phone: +92-48-923-0811-15 ext. 609 (Received 5th Jan 2019; accepted 26th Feb 2019) Abstract. This paper presents the first quantitative ethnobotanical knowledge and practices of using native plants for different ailments from Namal Valley of Pakistan. Data was gathered by interviewing 350 informants through semi-structured questionnaires. A total of 217 taxa belonging to 166 genera and 70 families were documented. Fabaceae and Asteraceae families were found to be the most cited families (with 19 and 18 species receptively). Herbs represent the most cited life form (71%) and flower was the most widely used part (34.8%) with decoction as main mode of the utilization (41.5%). On the basis of use values, the most commonly used ethnobotanical taxa in the Valley were reported to be Euphorbia heterophylla (0.7) and Merremia dissecta (0.6). The highest RFC value was noted for Aloe vera (0.14) while highest ICF value was estimated for dental problems category (0.7). -

Checklist of the Vascular Plants of Redwood National Park
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 9-17-2018 Checklist of the Vascular Plants of Redwood National Park James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Checklist of the Vascular Plants of Redwood National Park" (2018). Botanical Studies. 85. https://digitalcommons.humboldt.edu/botany_jps/85 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. A CHECKLIST OF THE VASCULAR PLANTS OF THE REDWOOD NATIONAL & STATE PARKS James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State Univerity Arcata, California 14 September 2018 The Redwood National and State Parks are located in Del Norte and Humboldt counties in coastal northwestern California. The national park was F E R N S established in 1968. In 1994, a cooperative agreement with the California Department of Parks and Recreation added Del Norte Coast, Prairie Creek, Athyriaceae – Lady Fern Family and Jedediah Smith Redwoods state parks to form a single administrative Athyrium filix-femina var. cyclosporum • northwestern lady fern unit. Together they comprise about 133,000 acres (540 km2), including 37 miles of coast line. Almost half of the remaining old growth redwood forests Blechnaceae – Deer Fern Family are protected in these four parks. -

Towards an Updated Checklist of the Libyan Flora
Towards an updated checklist of the Libyan flora Article Published Version Creative Commons: Attribution 3.0 (CC-BY) Open access Gawhari, A. M. H., Jury, S. L. and Culham, A. (2018) Towards an updated checklist of the Libyan flora. Phytotaxa, 338 (1). pp. 1-16. ISSN 1179-3155 doi: https://doi.org/10.11646/phytotaxa.338.1.1 Available at http://centaur.reading.ac.uk/76559/ It is advisable to refer to the publisher’s version if you intend to cite from the work. See Guidance on citing . Published version at: http://dx.doi.org/10.11646/phytotaxa.338.1.1 Identification Number/DOI: https://doi.org/10.11646/phytotaxa.338.1.1 <https://doi.org/10.11646/phytotaxa.338.1.1> Publisher: Magnolia Press All outputs in CentAUR are protected by Intellectual Property Rights law, including copyright law. Copyright and IPR is retained by the creators or other copyright holders. Terms and conditions for use of this material are defined in the End User Agreement . www.reading.ac.uk/centaur CentAUR Central Archive at the University of Reading Reading’s research outputs online Phytotaxa 338 (1): 001–016 ISSN 1179-3155 (print edition) http://www.mapress.com/j/pt/ PHYTOTAXA Copyright © 2018 Magnolia Press Article ISSN 1179-3163 (online edition) https://doi.org/10.11646/phytotaxa.338.1.1 Towards an updated checklist of the Libyan flora AHMED M. H. GAWHARI1, 2, STEPHEN L. JURY 2 & ALASTAIR CULHAM 2 1 Botany Department, Cyrenaica Herbarium, Faculty of Sciences, University of Benghazi, Benghazi, Libya E-mail: [email protected] 2 University of Reading Herbarium, The Harborne Building, School of Biological Sciences, University of Reading, Whiteknights, Read- ing, RG6 6AS, U.K. -

Linum Lewisii (Lewis's Blue Flax) (Pdf)
Linum lewisii Lewis’s Blue Flax by Kathy Lloyd Montana Native Plant Society Photo: Drake Barton Linum lewisii (Lewis’s Blue Flax) ewis’s blue flax, Linum lewisii, was col- wether Lewis by Frederick Pursh, who described the lected on July 9, 1806 in Montana. It is plant in his 1814 Flora Americae Septentrionalis or not known for certain who collected the Flora of North America. Pursh attached a label to specimen that is housed in the Lewis & the specimen that reads, “Perennial Flax. Valleys of LClark Her barium in Philadelphia. It could have been the Rocky mountains. July 9th 1806.” This Lewis Meriwether Lewis, who was traveling along the Sun and Clark specimen is one of a group that was feared River from Lewis & Clark County into Cascade lost but was fortunately relocated in 1896 at the County, or William Clark, who was at Camp Fortu- American Philosophical Society and subsequently nate in Beaverhead County. Lewis’s journal entry placed on permanent loan to the Academy of Natural for the day says nothing of blue flax, but does talk Sciences. about the rain and cold, “we then proceeded and it Another sheet of Lewis’s blue flax is currently part rained without intermission wet us to the skin…the of the Lewis & Clark Herbarium, but present-day day continuing rainy and cold…” The preserved botanists question its authenticity as a Lewis and specimen in the Lewis & Clark Herbarium appears Clark collection. James Reveal, a preeminent bota- to be in good shape and has a partially opened nist at the University of Maryland, believes the flower. -
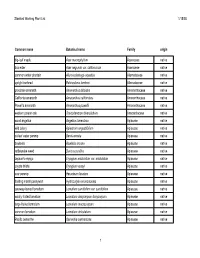
Plant List for Web Page
Stanford Working Plant List 1/15/08 Common name Botanical name Family origin big-leaf maple Acer macrophyllum Aceraceae native box elder Acer negundo var. californicum Aceraceae native common water plantain Alisma plantago-aquatica Alismataceae native upright burhead Echinodorus berteroi Alismataceae native prostrate amaranth Amaranthus blitoides Amaranthaceae native California amaranth Amaranthus californicus Amaranthaceae native Powell's amaranth Amaranthus powellii Amaranthaceae native western poison oak Toxicodendron diversilobum Anacardiaceae native wood angelica Angelica tomentosa Apiaceae native wild celery Apiastrum angustifolium Apiaceae native cutleaf water parsnip Berula erecta Apiaceae native bowlesia Bowlesia incana Apiaceae native rattlesnake weed Daucus pusillus Apiaceae native Jepson's eryngo Eryngium aristulatum var. aristulatum Apiaceae native coyote thistle Eryngium vaseyi Apiaceae native cow parsnip Heracleum lanatum Apiaceae native floating marsh pennywort Hydrocotyle ranunculoides Apiaceae native caraway-leaved lomatium Lomatium caruifolium var. caruifolium Apiaceae native woolly-fruited lomatium Lomatium dasycarpum dasycarpum Apiaceae native large-fruited lomatium Lomatium macrocarpum Apiaceae native common lomatium Lomatium utriculatum Apiaceae native Pacific oenanthe Oenanthe sarmentosa Apiaceae native 1 Stanford Working Plant List 1/15/08 wood sweet cicely Osmorhiza berteroi Apiaceae native mountain sweet cicely Osmorhiza chilensis Apiaceae native Gairdner's yampah (List 4) Perideridia gairdneri gairdneri Apiaceae -

Molecular Genetic Studies in Flax (Linum Usitatissimum L.)
Molecular genetic studies in flax (Linum usitatissimum L.) Moleculair genetische studies in vlas (Linum usitatissimum L.) Promotor: Prof. dr. ir. P. Stam Hoogleraar in de Plantenveredeling Copromotor: Dr. ir. H.J. van Eck Universitair docent, Laboratorium voor plantenveredeling Promotiecommissie: Prof. dr. A.G.M. Gerats (Radboud Universiteit Nijmegen) Prof. dr. R.F. Hoekstra (Wageningen Universiteit) Dr. R.G. van den Berg (Wageningen Universiteit) Dr. ir. J.W. van Ooijen (Kyazma B.V., Wageningen) Dit onderzoek is uitgevoerd binnen de onderzoeksscholen ‘Experimental Plant Sciences’ en ‘Production Ecology and Resource Conservation’. Molecular genetic studies in flax (Linum usitatissimum L.) Jaap Vromans Proefschrift ter verkrijging van de graad van doctor op gezag van de rector magnificus van Wageningen Universiteit, Prof. Dr. M.J. Kropff, in het openbaar te verdedigen op woensdag 15 maart 2006 des namiddags te vier uur in de Aula CIP-DTA Koninklijke Bibliotheek, Den Haag Molecular genetic studies in flax (Linum usitatissimum L.) Jaap Vromans PhD thesis, Wageningen University, The Netherlands With references – with summaries in Dutch and English ISBN 90-8504-374-3 Contents Chapter 1 General introduction 7 Chapter 2 Molecular analysis of flax (Linum usitatissimum L.) reveals a narrow genetic basis of fiber flax 21 Chapter 3 The molecular genetic variation in the genus Linum 41 Chapter 4 Selection of highly polymorphic AFLP fingerprints in flax (Linum usitatissimum L.) reduces the workload in genetic linkage map construction and results in -

Rigid Flax Linum Medium (Planch.) Britt
Rigid Flax Linum medium (Planch.) Britt. var. texanum (Planch.) Fern State Status: Threatened Federal Status: None Description: Rigid Flax is a perennial herb of the flax family (Linaceae), with yellow five-petaled flowers borne on stiff, ascending branches. Plants grow 2 to 7 dm (~8– 28 in.) in height. The flower petals are 4 to 8 mm long. The styles are distinct (i.e., not united at the base). The sepals are imbricate, and the inner ones have teeth with bulbous glandular tips along their edges. Leaves are entire, lance-shaped, and up to 2.5 cm (1 in.) long with the largest leaves towards the base of the plant. The upper leaves are alternate and usually have pointed tips, while those of the lowest nodes are opposite and blunt tipped. The sepals persist long after the petals have withered and subtend the small (2 mm), dry seed capsules. The species is most often found growing in barren, disturbed areas on sterile soil. Aids to identification: • Plants with stiffly ascending branches • Densely leaved with 30 to 70 leaves below the inflorescence • Lowest leaves opposite; upper leaves alternate • Seed capsules more-or-less spherical with a flattened top • Inner sepals with glandular teeth • Most easily identified when fruit are present Similar species: Four yellow-flowered Linum species that might be mistaken for Rigid Flax occur in Massachusetts. Grooved Yellow Flax (L. sulcatum var. sulcatum) differs from the other three in that it is an annual and its styles are united at the base. Woodland Yellow Flax (L. virginianum) and Panicled Yellow Flax (L. -

The History of Flax (Common Flax; Linum Usitatissimum)
The History of Flax (Common Flax; Linum usitatissimum) Ida Dyment http://www.flaxminnesota.com/Flax_Field_1_Med.jpg Common Flax (Linum usitatissimum) • Genus Linum, in Linaceae family. • Annual plant – grows up to 47 inches – thin leaves and stems. – Five-petal blue (to purple) flowers. http://www.mdidea.com/products/new/flax_flower01.jpg http://www.fleurdandeol.com/pic/msc/flax3.jpg The origins of Flax • Originated in the Mediterranean region into India. • Used in Egypt to wrap mummies • Early colonists grew small home plots • Began to be commercially processed starting around 1750 • Declined when cotton gin became popular Agriculture and Production • Flax grows well in temperate and sub-tropical regions in both hemisphere – France, Belgium, Russia, China, and Egypt grow a lot of the fiber flax currently – Minnesota and the Dakotas grow seed flax in the US • There are two common types of flax planted http://www.naturallygreen.co.uk/images/flaxseed.jpg Uses • Fiber – From Egypt to Modern Industry • Textiles, rope, paper • Food – Human food from seeds, fiber, oil – Livestock feed • Pharmacy – (Omega-3 fatty acids) – Heart health – skin treatment and cosmetics • Also used for ornamental purposes Omega-3 fatty acids • Omega-3 fatty acids in linseed oil may help prevent certain cancers. • Omega-3’s are also helpful for mental health • Helps lower cholesterol and regulate blood pressure • Flax also has very high level of lignans Many other uses • Flax in animal feed – Livestock and chickens • Aids digestion and • fortifies eggs • Flaxseed oil (linseed) also used for oil-painting agents, specialized paper, particleboard, varnish http://riverglenfarm.ca/__oneclick_uploads/2008/07/eggs.jpg http://www.sghongsheng.com/image/Plain%20particle%20board1.jpg http://www.frontera.com/assets/inline_images/39_braz%20cherry%20with%20linseed%20oil.jpg . -

SG Vol 10 1990.Pdf
ISSN 0394-9125 S'I'UDIA GEOBOTANICA An international journal Voi. 10 1990 EDITOR$ G. Estabrook - Ann Arbor, Mi L. lljanic - Zagreb E. Mayer - Ljubljana L. Orl6ci - London, On. F. Pedrotti - Camerino S. Pignatti - Roma A. Pirola - Pavia J. Poelt - Graz L. Poldini - Trieste E. Wikus Pignatti - Trieste EDITOR IN CHIEF D. Lausi · Trieste SECRETARY P.L. Nimis - Trieste Dipartimento di Biologia Sezione di Geobotanica ed Ecologia vegetale Università di Trieste STUDIA GEOBOTANICA 10: 3-13, 1990 SPATIAL PA'ITERNS AND DIVERSITY IN A POST-PLOUGHING SUCCESSION IN HIGH PLATEAU GRASSLANDS (PAMPA DE SAN LUIS, CORDOBA, ARGENTINA)* S. DiAZ, A. ACOSTA & M. CABIDO Keywords: Diversity, Grasslands, Ploughing, Spatial organization, Succession Abstract: Spatial arrangement and diversity along a post-ploughing succession are analyzed in a plateau at 1800 - 1900 m altitude in Centra! Argentina. Four successional stages were simultaneously studied: I (1 year of abandonment after ploughing), II (3-5 years), III (25 years) and IV (40 years). Detrended Correspondence Analysis and diversity analysis, comprising species diversity, spatial diversity and mean spatial niche width, were applied. It is concluded that (i) Spatial organization of grassland changes with succession: during early stages vegetation distribution follows a topographical gradient from upper to lower positions on slopes, whereas mosaic patterns prevail in late successional stages. (ii) As succession advances, species diversity increases, making us reject the hypothesis that species diversity decreases with succession as a result of the dominance of the grass Deyeuxia hieronymi. This process is associated with a progressive reduction of mean spatial niche width. Introduction The aim of this investigation was to describe the variability in spatial distribution of grasslands along a post-ploughing succession in the mountains of Central Argentina. -

Flax (Linum Usitatissimum L.) As a Model System
University of Alberta Environmental biosafety of genetically engineered crops: Flax (Linum usitatissimum L.) as a model system by Amitkumar Jayendrasinh Jhala A thesis submitted to the Faculty of Graduate Studies and Research in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Plant Science Department of Agricultural, Food and Nutritional Science ©Amitkumar Jayendrasinh Jhala Spring 2010 Edmonton, Alberta Permission is hereby granted to the University of Alberta Libraries to reproduce single copies of this thesis and to lend or sell such copies for private, scholarly or scientific research purposes only. Where the thesis is converted to, or otherwise made available in digital form, the University of Alberta will advise potential users of the thesis of these terms. The author reserves all other publication and other rights in association with the copyright in the thesis and, except as herein before provided, neither the thesis nor any substantial portion thereof may be printed or otherwise reproduced in any material form whatsoever without the author's prior written permission. Examining Committee Dr. Linda Hall, Agricultural, Food and Nutritional Science Dr. Randall Weselake, Agricultural, Food and Nutritional Science Dr. Lloyd Dosdall, Agricultural, Food and Nutritional Science Dr. Jocelyn Hall, Biological Sciences Dr. Robert Blackshaw, Agriculture and Agri-Food Canada, Lethbridge Dedicated to my Madam for her continuous love, support and encourangement Abstract Flax (Linum usitatissimum L.) is considered as a model plant species for multipurpose uses with whole plant utilization for several purposes including industril, food, animal feed, fiber, nutraceutical, pharmaceutical, and bioproduct markets. Therefore, flax is in the process of genetic engineering to meet the market requirements. -

Full Text (PDF)
Environmental and Experimental Biology (2015) 13: 123–131 Original Paper Use of anatomical characteristics for taxonomical study of some Iranian Linum taxa Seyed Mehdi Talebi1*, Maryam Rashnou-Taei2, Masoud Sheidai2, Zahra Noormohammadi3 1Department of Biology, Faculty of Sciences, Arak University, Arak, 38156-8-8349 Iran 2Faculty of Biological Sciences, Shahid Beheshti University, Tehran, Iran 3Department of Biology, School of Sciences, Tehran Science and Research Branch, Islamic Azad University, Tehran, Iran *Corresponding author, E-mail: [email protected] Abstract Linum is considered as the largest genus of Linaceae family, containing more than 180 species. The wide range of diversity within the genus Linum continues to challenge its taxonomical investigations. In present study anatomical features of vegetative organs, stem and leaf, of fourteen species, subspecies or varieties of three sections, were described with aim to improve the infrageneric classification of this genus. Plant samples were collected from natural populations of the studied taxa during 2011–2012. Embedded materials were used for microscopic investigation. Transverse hand sections of the lamina and stem were made from the middle part of fully-grown leaves and stems. Forty five qualitative and quantitative anatomical characteristics were examined in both stem and leaf. Principal Correspondence Analysis (PCA) of stem and leaf anatomical traits showed that some of these features were the most variable traits among the studied taxa. Analysis of variance showed