Genome-Wide Identification, Characterization, and Expression
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Protein Family Review
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by PubMed Central Protein family review The aquaporins comment Elisabeth Kruse, Norbert Uehlein and Ralf Kaldenhoff Address: Institute of Botany, Department of Applied Plant Sciences, Darmstadt University of Technology, Schnittspahnstraße 10, D-64287 Darmstadt, Germany. Correspondence: Ralf Kaldenhoff. Email: [email protected] reviews Published: 28 February 2006 Genome Biology 2006, 7:206 (doi:10.1186/gb-2006-7-2-206) The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2006/7/2/206 © 2006 BioMed Central Ltd reports Summary Water is the major component of all living cells, and efficient regulation of water homeostasis is essential for many biological processes. The mechanism by which water passes through biological membranes was a matter of debate until the discovery of the aquaporin water channels. Aquaporins are intrinsic membrane proteins characterized by six transmembrane helices that deposited research selectively allow water or other small uncharged molecules to pass along the osmotic gradient. In addition, recent observations show that some aquaporins also facilitate the transport of volatile substances, such as carbon dioxide (CO2) and ammonia (NH3), across membranes. Aquaporins usually form tetramers, with each monomer defining a single pore. Aquaporin-related proteins are found in all organisms, from archaea to mammals. In both uni- and multicellular organisms, numerous isoforms have been identified that are differentially expressed and modified by post- translational processes, thus allowing fine-tuned tissue-specific osmoregulation. In mammals, aquaporins are involved in multiple physiological processes, including kidney and salivary gland refereed research function. -

Single Amino Acid Substitutions in the Selectivity Filter
Ampah-Korsah et al. BMC Plant Biology (2017) 17:61 DOI 10.1186/s12870-017-1009-3 RESEARCHARTICLE Open Access Single amino acid substitutions in the selectivity filter render NbXIP1;1α aquaporin water permeable Henry Ampah-Korsah, Yonathan Sonntag, Angelica Engfors, Andreas Kirscht, Per Kjellbom and Urban Johanson* Abstract Background: Aquaporins (AQPs) are integral membrane proteins that facilitate transport of water and/or other small neutral solutes across membranes in all forms of life. The X Intrinsic Proteins (XIPs) are the most recently recognized and the least characterized aquaporin subfamily in higher plants. XIP1s have been shown to be impermeable to water but permeable to boric acid, glycerol, hydrogen peroxide and urea. However, uncertainty regarding the determinants for selectivity and lack of an activity that is easy to quantify have hindered functional investigations. In an effort to resolve these issues, we set out to introduce water permeability in Nicotiana benthamiana XIP1;1α (NbXIP1;1α), by exchanging amino acid residues of predicted alternative aromatic/arginine (ar/R) selectivity filters of NbXIP1;1α for residues constituting the water permeable ar/R selectivity filter of AtTIP2;1. Results: Here, we present functional results regarding the amino acid substitutions in the putative filters as well as deletions in loops C and D of NbXIP1;1α. In addition, homology models were created based on the high resolution X-ray structure of AtTIP2;1 to rationalize the functional properties of wild-type and mutant NbXIP1;1α. Our results favour Thr 246 rather than Val 242 as the residue at the helix 5 position in the ar/R filter of NbXIP1;1α and indicate that the pore is not occluded by the loops when heterologously expressed in Pichia pastoris. -

Combining Genome-Wide and Transcriptome-Wide Analyses Reveal
Li et al. BMC Genomics (2019) 20:538 https://doi.org/10.1186/s12864-019-5928-2 RESEARCHARTICLE Open Access Combining genome-wide and transcriptome-wide analyses reveal the evolutionary conservation and functional diversity of aquaporins in cotton Weixi Li, Dayong Zhang, Guozhong Zhu, Xinyue Mi and Wangzhen Guo* Abstract Background: Aquaporins (AQPs) are integral membrane proteins from a larger family of major intrinsic proteins (MIPs) and function in a huge variety of processes such as water transport, plant growth and stress response. The availability of the whole-genome data of different cotton species allows us to study systematic evolution and function of cotton AQPs on a genome-wide level. Results: Here, a total of 53, 58, 113 and 111 AQP genes were identified in G. arboreum, G. raimondii, G. hirsutum and G. barbadense, respectively. A comprehensive analysis of cotton AQPs, involved in exon/intron structure, functional domains, phylogenetic relationships and gene duplications, divided these AQPs into five subfamilies (PIP, NIP, SIP, TIP and XIP). Comparative genome analysis among 30 species from algae to angiosperm as well as common tandem duplication events in 24 well-studied plants further revealed the evolutionary conservation of AQP family in the organism kingdom. Combining transcriptome analysis and Quantitative Real-time PCR (qRT-PCR) verification, most AQPs exhibited tissue-specific expression patterns both in G. raimondii and G. hirsutum. Meanwhile, a bias of time to peak expression of several AQPs was also detected after treating G. davidsonii and G. hirsutum with 200 mM NaCl. It is interesting that both PIP1;4 h/i/j and PIP2;2a/e showed the highly conserved tandem structure, but differentially contributed to tissue development and stress response in different cotton species. -

Anti-HIV-1 Activities of the Extracts from the Medicinal Plant Linum Grandiflorum Desf
® Medicinal and Aromatic Plant Science and Biotechnology ©2009 Global Science Books Anti-HIV-1 Activities of the Extracts from the Medicinal Plant Linum grandiflorum Desf. Magdy M. D. Mohammed1,2,3* • Lars P. Christensen1,4 • Nabaweya A. Ibrahim3 • Nagwa E. Awad3 • Ibrahim F. Zeid5 • Erik B. Pedersen2 • Kenneth B. Jensen2 • Paolo L. Colla6 1 Danish Institute of Agricultural Sciences (DIAS), Department of Food Science, Research Center Aarslev, Aarhus University, Kirstinebjergvej 10, DK-5792 Aarslev, Denmark 2 Nucleic Acid Center, Institute of Physics and Chemistry, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark 3 Pharmacognosy Department, National Research Center, Dokki-12311, Cairo, Egypt 4 Institute of Chemical Engineering, Biotechnology and Environmental Technology, University of Southern Denmark, Niels Bohrs Allé 1, DK-5230 Odense M, Denmark 5 Chemistry Department, Faculty of Science, El-Menoufia University, El-Menoufia, Egypt 6 Department of Science and Biomedical Technology, Cittadella University, Monserrato, Italy Corresponding author : * [email protected] ABSTRACT As part of our screening of anti-AIDS agents from natural sources e.g. Ixora undulata, Paulownia tomentosa, Fortunella margarita, Aegle marmelos and Erythrina abyssinica, the different organic and aqueous extracts of Linum grandiflorum leaves and seeds were evaluated in vitro by the microculture tetrazolium (MTT) assay. The activity of the tested extracts against multiplication of HIV-1 wild type IIIB, N119, A17, and EFVR in acutely infected cells was based on inhibition of virus-induced cytopathicity in MT-4 cells. Results revealed that both the MeOH and the CHCl3 extracts of L. grandiflorum have significant inhibitory effects against HIV-1 induced infection with MT-4 cells. -

Checklist of the Vascular Plants of Redwood National Park
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 9-17-2018 Checklist of the Vascular Plants of Redwood National Park James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Checklist of the Vascular Plants of Redwood National Park" (2018). Botanical Studies. 85. https://digitalcommons.humboldt.edu/botany_jps/85 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. A CHECKLIST OF THE VASCULAR PLANTS OF THE REDWOOD NATIONAL & STATE PARKS James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State Univerity Arcata, California 14 September 2018 The Redwood National and State Parks are located in Del Norte and Humboldt counties in coastal northwestern California. The national park was F E R N S established in 1968. In 1994, a cooperative agreement with the California Department of Parks and Recreation added Del Norte Coast, Prairie Creek, Athyriaceae – Lady Fern Family and Jedediah Smith Redwoods state parks to form a single administrative Athyrium filix-femina var. cyclosporum • northwestern lady fern unit. Together they comprise about 133,000 acres (540 km2), including 37 miles of coast line. Almost half of the remaining old growth redwood forests Blechnaceae – Deer Fern Family are protected in these four parks. -
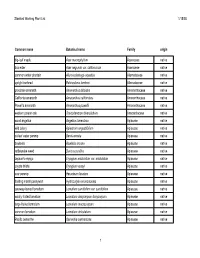
Plant List for Web Page
Stanford Working Plant List 1/15/08 Common name Botanical name Family origin big-leaf maple Acer macrophyllum Aceraceae native box elder Acer negundo var. californicum Aceraceae native common water plantain Alisma plantago-aquatica Alismataceae native upright burhead Echinodorus berteroi Alismataceae native prostrate amaranth Amaranthus blitoides Amaranthaceae native California amaranth Amaranthus californicus Amaranthaceae native Powell's amaranth Amaranthus powellii Amaranthaceae native western poison oak Toxicodendron diversilobum Anacardiaceae native wood angelica Angelica tomentosa Apiaceae native wild celery Apiastrum angustifolium Apiaceae native cutleaf water parsnip Berula erecta Apiaceae native bowlesia Bowlesia incana Apiaceae native rattlesnake weed Daucus pusillus Apiaceae native Jepson's eryngo Eryngium aristulatum var. aristulatum Apiaceae native coyote thistle Eryngium vaseyi Apiaceae native cow parsnip Heracleum lanatum Apiaceae native floating marsh pennywort Hydrocotyle ranunculoides Apiaceae native caraway-leaved lomatium Lomatium caruifolium var. caruifolium Apiaceae native woolly-fruited lomatium Lomatium dasycarpum dasycarpum Apiaceae native large-fruited lomatium Lomatium macrocarpum Apiaceae native common lomatium Lomatium utriculatum Apiaceae native Pacific oenanthe Oenanthe sarmentosa Apiaceae native 1 Stanford Working Plant List 1/15/08 wood sweet cicely Osmorhiza berteroi Apiaceae native mountain sweet cicely Osmorhiza chilensis Apiaceae native Gairdner's yampah (List 4) Perideridia gairdneri gairdneri Apiaceae -

Molecular Genetic Studies in Flax (Linum Usitatissimum L.)
Molecular genetic studies in flax (Linum usitatissimum L.) Moleculair genetische studies in vlas (Linum usitatissimum L.) Promotor: Prof. dr. ir. P. Stam Hoogleraar in de Plantenveredeling Copromotor: Dr. ir. H.J. van Eck Universitair docent, Laboratorium voor plantenveredeling Promotiecommissie: Prof. dr. A.G.M. Gerats (Radboud Universiteit Nijmegen) Prof. dr. R.F. Hoekstra (Wageningen Universiteit) Dr. R.G. van den Berg (Wageningen Universiteit) Dr. ir. J.W. van Ooijen (Kyazma B.V., Wageningen) Dit onderzoek is uitgevoerd binnen de onderzoeksscholen ‘Experimental Plant Sciences’ en ‘Production Ecology and Resource Conservation’. Molecular genetic studies in flax (Linum usitatissimum L.) Jaap Vromans Proefschrift ter verkrijging van de graad van doctor op gezag van de rector magnificus van Wageningen Universiteit, Prof. Dr. M.J. Kropff, in het openbaar te verdedigen op woensdag 15 maart 2006 des namiddags te vier uur in de Aula CIP-DTA Koninklijke Bibliotheek, Den Haag Molecular genetic studies in flax (Linum usitatissimum L.) Jaap Vromans PhD thesis, Wageningen University, The Netherlands With references – with summaries in Dutch and English ISBN 90-8504-374-3 Contents Chapter 1 General introduction 7 Chapter 2 Molecular analysis of flax (Linum usitatissimum L.) reveals a narrow genetic basis of fiber flax 21 Chapter 3 The molecular genetic variation in the genus Linum 41 Chapter 4 Selection of highly polymorphic AFLP fingerprints in flax (Linum usitatissimum L.) reduces the workload in genetic linkage map construction and results in -

Illustrated Flora of East Texas Illustrated Flora of East Texas
ILLUSTRATED FLORA OF EAST TEXAS ILLUSTRATED FLORA OF EAST TEXAS IS PUBLISHED WITH THE SUPPORT OF: MAJOR BENEFACTORS: DAVID GIBSON AND WILL CRENSHAW DISCOVERY FUND U.S. FISH AND WILDLIFE FOUNDATION (NATIONAL PARK SERVICE, USDA FOREST SERVICE) TEXAS PARKS AND WILDLIFE DEPARTMENT SCOTT AND STUART GENTLING BENEFACTORS: NEW DOROTHEA L. LEONHARDT FOUNDATION (ANDREA C. HARKINS) TEMPLE-INLAND FOUNDATION SUMMERLEE FOUNDATION AMON G. CARTER FOUNDATION ROBERT J. O’KENNON PEG & BEN KEITH DORA & GORDON SYLVESTER DAVID & SUE NIVENS NATIVE PLANT SOCIETY OF TEXAS DAVID & MARGARET BAMBERGER GORDON MAY & KAREN WILLIAMSON JACOB & TERESE HERSHEY FOUNDATION INSTITUTIONAL SUPPORT: AUSTIN COLLEGE BOTANICAL RESEARCH INSTITUTE OF TEXAS SID RICHARDSON CAREER DEVELOPMENT FUND OF AUSTIN COLLEGE II OTHER CONTRIBUTORS: ALLDREDGE, LINDA & JACK HOLLEMAN, W.B. PETRUS, ELAINE J. BATTERBAE, SUSAN ROBERTS HOLT, JEAN & DUNCAN PRITCHETT, MARY H. BECK, NELL HUBER, MARY MAUD PRICE, DIANE BECKELMAN, SARA HUDSON, JIM & YONIE PRUESS, WARREN W. BENDER, LYNNE HULTMARK, GORDON & SARAH ROACH, ELIZABETH M. & ALLEN BIBB, NATHAN & BETTIE HUSTON, MELIA ROEBUCK, RICK & VICKI BOSWORTH, TONY JACOBS, BONNIE & LOUIS ROGNLIE, GLORIA & ERIC BOTTONE, LAURA BURKS JAMES, ROI & DEANNA ROUSH, LUCY BROWN, LARRY E. JEFFORDS, RUSSELL M. ROWE, BRIAN BRUSER, III, MR. & MRS. HENRY JOHN, SUE & PHIL ROZELL, JIMMY BURT, HELEN W. JONES, MARY LOU SANDLIN, MIKE CAMPBELL, KATHERINE & CHARLES KAHLE, GAIL SANDLIN, MR. & MRS. WILLIAM CARR, WILLIAM R. KARGES, JOANN SATTERWHITE, BEN CLARY, KAREN KEITH, ELIZABETH & ERIC SCHOENFELD, CARL COCHRAN, JOYCE LANEY, ELEANOR W. SCHULTZE, BETTY DAHLBERG, WALTER G. LAUGHLIN, DR. JAMES E. SCHULZE, PETER & HELEN DALLAS CHAPTER-NPSOT LECHE, BEVERLY SENNHAUSER, KELLY S. DAMEWOOD, LOGAN & ELEANOR LEWIS, PATRICIA SERLING, STEVEN DAMUTH, STEVEN LIGGIO, JOE SHANNON, LEILA HOUSEMAN DAVIS, ELLEN D. -

Flora Mediterranea 26
FLORA MEDITERRANEA 26 Published under the auspices of OPTIMA by the Herbarium Mediterraneum Panormitanum Palermo – 2016 FLORA MEDITERRANEA Edited on behalf of the International Foundation pro Herbario Mediterraneo by Francesco M. Raimondo, Werner Greuter & Gianniantonio Domina Editorial board G. Domina (Palermo), F. Garbari (Pisa), W. Greuter (Berlin), S. L. Jury (Reading), G. Kamari (Patras), P. Mazzola (Palermo), S. Pignatti (Roma), F. M. Raimondo (Palermo), C. Salmeri (Palermo), B. Valdés (Sevilla), G. Venturella (Palermo). Advisory Committee P. V. Arrigoni (Firenze) P. Küpfer (Neuchatel) H. M. Burdet (Genève) J. Mathez (Montpellier) A. Carapezza (Palermo) G. Moggi (Firenze) C. D. K. Cook (Zurich) E. Nardi (Firenze) R. Courtecuisse (Lille) P. L. Nimis (Trieste) V. Demoulin (Liège) D. Phitos (Patras) F. Ehrendorfer (Wien) L. Poldini (Trieste) M. Erben (Munchen) R. M. Ros Espín (Murcia) G. Giaccone (Catania) A. Strid (Copenhagen) V. H. Heywood (Reading) B. Zimmer (Berlin) Editorial Office Editorial assistance: A. M. Mannino Editorial secretariat: V. Spadaro & P. Campisi Layout & Tecnical editing: E. Di Gristina & F. La Sorte Design: V. Magro & L. C. Raimondo Redazione di "Flora Mediterranea" Herbarium Mediterraneum Panormitanum, Università di Palermo Via Lincoln, 2 I-90133 Palermo, Italy [email protected] Printed by Luxograph s.r.l., Piazza Bartolomeo da Messina, 2/E - Palermo Registration at Tribunale di Palermo, no. 27 of 12 July 1991 ISSN: 1120-4052 printed, 2240-4538 online DOI: 10.7320/FlMedit26.001 Copyright © by International Foundation pro Herbario Mediterraneo, Palermo Contents V. Hugonnot & L. Chavoutier: A modern record of one of the rarest European mosses, Ptychomitrium incurvum (Ptychomitriaceae), in Eastern Pyrenees, France . 5 P. Chène, M. -

How Does Genome Size Affect the Evolution of Pollen Tube Growth Rate, a Haploid Performance Trait?
Manuscript bioRxiv preprint doi: https://doi.org/10.1101/462663; this version postedClick April here18, 2019. to The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv aaccess/download;Manuscript;PTGR.genome.evolution.15April20 license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Effects of genome size on pollen performance 2 3 4 5 How does genome size affect the evolution of pollen tube growth rate, a haploid 6 performance trait? 7 8 9 10 11 John B. Reese1,2 and Joseph H. Williams2 12 Department of Ecology and Evolutionary Biology, University of Tennessee, Knoxville, TN 13 37996, U.S.A. 14 15 16 17 1Author for correspondence: 18 John B. Reese 19 Tel: 865 974 9371 20 Email: [email protected] 21 1 bioRxiv preprint doi: https://doi.org/10.1101/462663; this version posted April 18, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 22 ABSTRACT 23 Premise of the Study – Male gametophytes of most seed plants deliver sperm to eggs via a 24 pollen tube. Pollen tube growth rates (PTGRs) of angiosperms are exceptionally rapid, a pattern 25 attributed to more effective haploid selection under stronger pollen competition. Paradoxically, 26 whole genome duplication (WGD) has been common in angiosperms but rare in gymnosperms. -

How Does Genome Size Affect the Evolution of Pollen Tube Growth Rate, a Haploid Performance Trait?
bioRxiv preprint doi: https://doi.org/10.1101/462663; this version posted November 5, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Effects of genome size on pollen performance 2 3 4 5 6 How does genome size affect the evolution of pollen tube growth rate, a haploid 7 performance trait? 8 9 10 11 12 John B. Reese1,2 and Joseph H. Williams1 13 Department of Ecology and Evolutionary Biology, University of Tennessee, Knoxville, TN 14 37996, U.S.A. 15 16 17 18 1Author for correspondence: 19 John B. Reese 20 Tel: 865 974 9371 21 Email: [email protected] 22 23 24 1 bioRxiv preprint doi: https://doi.org/10.1101/462663; this version posted November 5, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 25 ABSTRACT 26 Premise of the Study - Male gametophytes of seed plants deliver sperm to eggs via a pollen 27 tube. Pollen tube growth rate (PTGR) may evolve rapidly due to pollen competition and haploid 28 selection, but many angiosperms are currently polyploid and all have polyploid histories. 29 Polyploidy should initially accelerate PTGR via “genotypic effects” of increased gene dosage 30 and heterozygosity on metabolic rates, but “nucleotypic effects” of genome size on cell size 31 should reduce PTGR. -

Flax (Linum Usitatissimum L.) As a Model System
University of Alberta Environmental biosafety of genetically engineered crops: Flax (Linum usitatissimum L.) as a model system by Amitkumar Jayendrasinh Jhala A thesis submitted to the Faculty of Graduate Studies and Research in partial fulfillment of the requirements for the degree of Doctor of Philosophy in Plant Science Department of Agricultural, Food and Nutritional Science ©Amitkumar Jayendrasinh Jhala Spring 2010 Edmonton, Alberta Permission is hereby granted to the University of Alberta Libraries to reproduce single copies of this thesis and to lend or sell such copies for private, scholarly or scientific research purposes only. Where the thesis is converted to, or otherwise made available in digital form, the University of Alberta will advise potential users of the thesis of these terms. The author reserves all other publication and other rights in association with the copyright in the thesis and, except as herein before provided, neither the thesis nor any substantial portion thereof may be printed or otherwise reproduced in any material form whatsoever without the author's prior written permission. Examining Committee Dr. Linda Hall, Agricultural, Food and Nutritional Science Dr. Randall Weselake, Agricultural, Food and Nutritional Science Dr. Lloyd Dosdall, Agricultural, Food and Nutritional Science Dr. Jocelyn Hall, Biological Sciences Dr. Robert Blackshaw, Agriculture and Agri-Food Canada, Lethbridge Dedicated to my Madam for her continuous love, support and encourangement Abstract Flax (Linum usitatissimum L.) is considered as a model plant species for multipurpose uses with whole plant utilization for several purposes including industril, food, animal feed, fiber, nutraceutical, pharmaceutical, and bioproduct markets. Therefore, flax is in the process of genetic engineering to meet the market requirements.