APC Anti-Human Cd11b (ICRF44) Catalog Number: 20-0118
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Propranolol-Mediated Attenuation of MMP-9 Excretion in Infants with Hemangiomas
Supplementary Online Content Thaivalappil S, Bauman N, Saieg A, Movius E, Brown KJ, Preciado D. Propranolol-mediated attenuation of MMP-9 excretion in infants with hemangiomas. JAMA Otolaryngol Head Neck Surg. doi:10.1001/jamaoto.2013.4773 eTable. List of All of the Proteins Identified by Proteomics This supplementary material has been provided by the authors to give readers additional information about their work. © 2013 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 10/01/2021 eTable. List of All of the Proteins Identified by Proteomics Protein Name Prop 12 mo/4 Pred 12 mo/4 Δ Prop to Pred mo mo Myeloperoxidase OS=Homo sapiens GN=MPO 26.00 143.00 ‐117.00 Lactotransferrin OS=Homo sapiens GN=LTF 114.00 205.50 ‐91.50 Matrix metalloproteinase‐9 OS=Homo sapiens GN=MMP9 5.00 36.00 ‐31.00 Neutrophil elastase OS=Homo sapiens GN=ELANE 24.00 48.00 ‐24.00 Bleomycin hydrolase OS=Homo sapiens GN=BLMH 3.00 25.00 ‐22.00 CAP7_HUMAN Azurocidin OS=Homo sapiens GN=AZU1 PE=1 SV=3 4.00 26.00 ‐22.00 S10A8_HUMAN Protein S100‐A8 OS=Homo sapiens GN=S100A8 PE=1 14.67 30.50 ‐15.83 SV=1 IL1F9_HUMAN Interleukin‐1 family member 9 OS=Homo sapiens 1.00 15.00 ‐14.00 GN=IL1F9 PE=1 SV=1 MUC5B_HUMAN Mucin‐5B OS=Homo sapiens GN=MUC5B PE=1 SV=3 2.00 14.00 ‐12.00 MUC4_HUMAN Mucin‐4 OS=Homo sapiens GN=MUC4 PE=1 SV=3 1.00 12.00 ‐11.00 HRG_HUMAN Histidine‐rich glycoprotein OS=Homo sapiens GN=HRG 1.00 12.00 ‐11.00 PE=1 SV=1 TKT_HUMAN Transketolase OS=Homo sapiens GN=TKT PE=1 SV=3 17.00 28.00 ‐11.00 CATG_HUMAN Cathepsin G OS=Homo -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

CD Markers Are Routinely Used for the Immunophenotyping of Cells
ptglab.com 1 CD MARKER ANTIBODIES www.ptglab.com Introduction The cluster of differentiation (abbreviated as CD) is a protocol used for the identification and investigation of cell surface molecules. So-called CD markers are routinely used for the immunophenotyping of cells. Despite this use, they are not limited to roles in the immune system and perform a variety of roles in cell differentiation, adhesion, migration, blood clotting, gamete fertilization, amino acid transport and apoptosis, among many others. As such, Proteintech’s mini catalog featuring its antibodies targeting CD markers is applicable to a wide range of research disciplines. PRODUCT FOCUS PECAM1 Platelet endothelial cell adhesion of blood vessels – making up a large portion molecule-1 (PECAM1), also known as cluster of its intracellular junctions. PECAM-1 is also CD Number of differentiation 31 (CD31), is a member of present on the surface of hematopoietic the immunoglobulin gene superfamily of cell cells and immune cells including platelets, CD31 adhesion molecules. It is highly expressed monocytes, neutrophils, natural killer cells, on the surface of the endothelium – the thin megakaryocytes and some types of T-cell. Catalog Number layer of endothelial cells lining the interior 11256-1-AP Type Rabbit Polyclonal Applications ELISA, FC, IF, IHC, IP, WB 16 Publications Immunohistochemical of paraffin-embedded Figure 1: Immunofluorescence staining human hepatocirrhosis using PECAM1, CD31 of PECAM1 (11256-1-AP), Alexa 488 goat antibody (11265-1-AP) at a dilution of 1:50 anti-rabbit (green), and smooth muscle KD/KO Validated (40x objective). alpha-actin (red), courtesy of Nicola Smart. PECAM1: Customer Testimonial Nicola Smart, a cardiovascular researcher “As you can see [the immunostaining] is and a group leader at the University of extremely clean and specific [and] displays Oxford, has said of the PECAM1 antibody strong intercellular junction expression, (11265-1-AP) that it “worked beautifully as expected for a cell adhesion molecule.” on every occasion I’ve tried it.” Proteintech thanks Dr. -
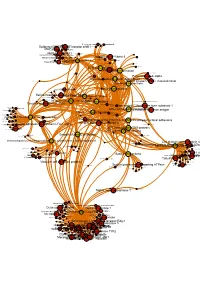
Brcaallwithlabelsintegrina1b1
Receptor-type tyrosine-proteinMuscle- skeletal receptor phosphatase tyrosine protein kinase S Dual specificity mitogen-activated protein kinase kinase 1 Plasminogen activator inhibitor-1 Epidermal growth factor receptor erbB1 Beta amyloid A4 protein MAPDual specificity Epidermalkinase mitogen-activated growth factorCasein proteinreceptor ERK2 kinase and ErbB2kinase; II (HER1 alphaMEK1/2 and (prime) HER2) Casein kinase II alpha Integrin alpha-IIb MAPCasein kinase kinase II beta ERK1 Dual specificity mitogen-activated protein kinase kinase 2 Lysyl oxidase Integrin alpha-IIb/beta-3 Furin Mitogen-activatedSerine/threonine-protein kinase RAF and Dual specificityEpidermal protein mitogen-activated growth proteinkinase; factor kinase kinase receptor 1 (Raf/MEK)ERK1/ERK2 Integrin alpha-V/beta-3 Integrin alpha-V/beta-3 and alpha-IIb/beta 3 Casein kinase II Signal transduction by L1 Integrin alpha-V/beta-6 Casein kinase II alpha/beta Vitronectin receptor alpha Bone morphogenetic protein 2 ECM proteoglycans Bone morphogenetic protein 4 VitronectinElastic fibre formation Peripheral plasma membrane protein CASK Neuropilin-1 Integrin alpha-V/beta-5 Protein kinaseProtein kinase C (PKC) alpha Integrin alpha-2/beta-3 Molecules associated with elastic fibres PKC alpha and beta-2 MER intracellular domain/EGFR extracellular domain chimera Fibronectin receptor alpha Protein kinase C- PKC; classical/novel Syndecan interactions Integrin alpha-2 von Willebrand factorIntegrin alpha-10 Laminin interactions Integrin alpha-11 Fibrinogen beta chain Serine/threonine-proteinVascular -

High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype
Supplementary Information High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype Håkon Reikvam 1,2,*, Elise Aasebø 1, Annette K. Brenner 2, Sushma Bartaula-Brevik 1, Ida Sofie Grønningsæter 2, Rakel Brendsdal Forthun 2, Randi Hovland 3,4 and Øystein Bruserud 1,2 1 Department of Clinical Science, University of Bergen, 5020, Bergen, Norway 2 Department of Medicine, Haukeland University Hospital, 5021, Bergen, Norway 3 Department of Medical Genetics, Haukeland University Hospital, 5021, Bergen, Norway 4 Institute of Biomedicine, University of Bergen, 5020, Bergen, Norway * Correspondence: [email protected]; Tel.: +55-97-50-00 J. Clin. Med. 2019, 8, x 2 of 36 Figure S1. Mutational studies in a cohort of 71 AML patients. The figure shows the number of patients with the various mutations (upper), the number of mutations in for each patient (middle) and the number of main classes with mutation(s) in each patient (lower). 2 J. Clin. Med. 2019, 8, x; doi: www.mdpi.com/journal/jcm J. Clin. Med. 2019, 8, x 3 of 36 Figure S2. The immunophenotype of primary human AML cells derived from 62 unselected patients. The expression of the eight differentiation markers CD13, CD14, CD15, CD33, CD34, CD45, CD117 and HLA-DR was investigated for 62 of the 71 patients included in our present study. We performed an unsupervised hierarchical cluster analysis and identified four patient main clusters/patient subsets. The mutational profile for each f the 62 patients is also given (middle), no individual mutation of main class of mutations showed any significant association with any of the for differentiation marker clusters (middle). -

Structural Insight on the Recognition of Surface-Bound Opsonins by the Integrin I Domain of Complement Receptor 3
Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3 Goran Bajica, Laure Yatimea, Robert B. Simb, Thomas Vorup-Jensenc,1, and Gregers R. Andersena,1 Departments of aMolecular Biology and Genetics and cBiomedicine, Aarhus University, DK-8000 Aarhus, Denmark; and bDepartment of Pharmacology, University of Oxford, Oxford OX1 3QT, United Kingdom Edited by Douglas T. Fearon, University of Cambridge School of Clinical Medicine, Cambridge, United Kingdom, and approved August 28, 2013 (received for review June 13, 2013) Complement receptors (CRs), expressed notably on myeloid and degranulation, and changes in leukocyte cytokine production (2, lymphoid cells, play an essential function in the elimination of 5–7). CR3, and to a lesser degree CR4, are essential for the complement-opsonized pathogens and apoptotic/necrotic cells. In phagocytosis of complement-opsonized particles or complexes addition, these receptors are crucial for the cross-talk between the (6, 8, 9). Complement-opsonized immune complexes are cap- innate andadaptive branches ofthe immune system. CR3 (also known tured in the lymph nodes by CR3-positive subcapsular sinus as Mac-1, integrin α β , or CD11b/CD18) is expressed on all macro- macrophages (SSMs) and conveyed directly to naïve B cells or M 2 γ phages and recognizes iC3b on complement-opsonized objects, en- through follicular dendritic cells (10) using CR1, CR2, and Fc abling their phagocytosis. We demonstrate that the C3d moiety of receptors for antigen capture (11, 12). Hence, antigen-presenting iC3b harbors the binding site for the CR3 αI domain, and our structure cells such as SSMs may act as antigen storage and provide B of the C3d:αI domain complex rationalizes the CR3 selectivity for iC3b. -

Structural Insight on the Recognition of Surface-Bound Opsonins by the Integrin I Domain of Complement Receptor 3
Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3 Goran Bajica, Laure Yatimea, Robert B. Simb, Thomas Vorup-Jensenc,1, and Gregers R. Andersena,1 Departments of aMolecular Biology and Genetics and cBiomedicine, Aarhus University, DK-8000 Aarhus, Denmark; and bDepartment of Pharmacology, University of Oxford, Oxford OX1 3QT, United Kingdom Edited by Douglas T. Fearon, University of Cambridge School of Clinical Medicine, Cambridge, United Kingdom, and approved August 28, 2013 (received for review June 13, 2013) Complement receptors (CRs), expressed notably on myeloid and degranulation, and changes in leukocyte cytokine production (2, lymphoid cells, play an essential function in the elimination of 5–7). CR3, and to a lesser degree CR4, are essential for the complement-opsonized pathogens and apoptotic/necrotic cells. In phagocytosis of complement-opsonized particles or complexes addition, these receptors are crucial for the cross-talk between the (6, 8, 9). Complement-opsonized immune complexes are cap- innate andadaptive branches ofthe immune system. CR3 (also known tured in the lymph nodes by CR3-positive subcapsular sinus as Mac-1, integrin α β , or CD11b/CD18) is expressed on all macro- macrophages (SSMs) and conveyed directly to naïve B cells or M 2 γ phages and recognizes iC3b on complement-opsonized objects, en- through follicular dendritic cells (10) using CR1, CR2, and Fc abling their phagocytosis. We demonstrate that the C3d moiety of receptors for antigen capture (11, 12). Hence, antigen-presenting iC3b harbors the binding site for the CR3 αI domain, and our structure cells such as SSMs may act as antigen storage and provide B of the C3d:αI domain complex rationalizes the CR3 selectivity for iC3b. -

Vitro Generated to Ex Vivo CD8 T Cells Comparative and Functional
Downloaded from http://www.jimmunol.org/ by guest on September 27, 2021 is online at: average * The Journal of Immunology , 21 of which you can access for free at: 2012; 189:3411-3420; Prepublished online 27 from submission to initial decision 4 weeks from acceptance to publication Comparative and Functional Evaluation of In Vitro Generated to Ex Vivo CD8 T Cells Dzana D. Dervovic, Maria Ciofani, Korosh Kianizad and Juan Carlos Zúñiga-Pflücker J Immunol August 2012; doi: 10.4049/jimmunol.1200979 http://www.jimmunol.org/content/189/7/3411 cites 61 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2012/08/28/jimmunol.120097 9.DC1 This article http://www.jimmunol.org/content/189/7/3411.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 27, 2021. The Journal of Immunology Comparative and Functional Evaluation of In Vitro Generated to Ex Vivo CD8 T Cells Dzˇana D. Dervovic´,*,† Maria Ciofani,*,†,1 Korosh Kianizad,*,† and Juan Carlos Zu´n˜iga-Pflu¨cker*,† The generation of the cytotoxic CD8 T cell response is dependent on the functional outcomes imposed by the intrathymic constraints of differentiation and self-tolerance. -

Datasheet: VMA00481 Product Details
Datasheet: VMA00481 Description: MOUSE ANTI CD11b Specificity: CD11b Format: Purified Product Type: PrecisionAb™ Monoclonal Clone: 2D11 Isotype: IgG1 Quantity: 100 µl Product Details Applications This product has been reported to work in the following applications. This information is derived from testing within our laboratories, peer-reviewed publications or personal communications from the originators. Please refer to references indicated for further information. For general protocol recommendations, please visit www.bio-rad-antibodies.com/protocols. Yes No Not Determined Suggested Dilution Western Blotting 1/1000 PrecisionAb antibodies have been extensively validated for the western blot application. The antibody has been validated at the suggested dilution. Where this product has not been tested for use in a particular technique this does not necessarily exclude its use in such procedures. Further optimization may be required dependant on sample type. Target Species Human Product Form Purified IgG - liquid Preparation Purified IgG prepared by affinity chromatography Buffer Solution Phosphate buffered saline Preservative 0.09% Sodium Azide (NaN3) Stabilisers 1% Bovine Serum Albumin 50% Glycerol Immunogen Full length recombinant protein of human CD11b produced in HEK293T cells External Database Links UniProt: P11215 Related reagents Entrez Gene: 3684 ITGAM Related reagents Synonyms CD11B, CR3A Page 1 of 2 Specificity Mouse anti Human CD11b antibody recognizes CD11b also known as integrin alpha-M, CD11 antigen-like family member B, CR-3 alpha chain, cell surface glycoprotein MAC-1 subunit alpha, leukocyte adhesion receptor MO1, macrophage antigen alpha polypeptide or neutrophil adherence receptor alpha-M subunit. The ITGAM gene encodes the integrin alpha M chain. Integrins are heterodimeric integral membrane proteins composed of an alpha chain and a beta chain. -

Cell Adhesion Molecules Are Mediated by Photobiomodulation at 660 Nm in Diabetic Wounded Fibroblast Cells
cells Article Cell Adhesion Molecules Are Mediated by Photobiomodulation at 660 nm in Diabetic Wounded Fibroblast Cells Nicolette N. Houreld * ID , Sandra M. Ayuk and Heidi Abrahamse ID Laser Research Centre, Faculty of Health Sciences, University of Johannesburg, P.O. Box 17011, Doornfontein, Johannesburg 2028, South Africa; [email protected] (S.M.A.); [email protected] (H.A.) * Correspondence: [email protected]; Tel.: +27-11-559-6833 Received: 9 March 2018; Accepted: 12 April 2018; Published: 16 April 2018 Abstract: Diabetes affects extracellular matrix (ECM) metabolism, contributing to delayed wound healing and lower limb amputation. Application of light (photobiomodulation, PBM) has been shown to improve wound healing. This study aimed to evaluate the influence of PBM on cell adhesion molecules (CAMs) in diabetic wound healing. Isolated human skin fibroblasts were grouped into a diabetic wounded model. A diode laser at 660 nm with a fluence of 5 J/cm2 was used for irradiation and cells were analysed 48 h post-irradiation. Controls consisted of sham-irradiated (0 J/cm2) cells. Real-time reverse transcription (RT) quantitative polymerase chain reaction (qPCR) was used to determine the expression of CAM-related genes. Ten genes were up-regulated in diabetic wounded cells, while 25 genes were down-regulated. Genes were related to transmembrane molecules, cell–cell adhesion, and cell–matrix adhesion, and also included genes related to other CAM molecules. PBM at 660 nm modulated gene expression of various CAMs contributing to the increased healing seen in clinical practice. There is a need for new therapies to improve diabetic wound healing. -

(MDR/TAP), Member 1 ABL1 NM 00
Official Symbol Accession Official Full Name ABCB1 NM_000927.3 ATP-binding cassette, sub-family B (MDR/TAP), member 1 ABL1 NM_005157.3 c-abl oncogene 1, non-receptor tyrosine kinase ADA NM_000022.2 adenosine deaminase AHR NM_001621.3 aryl hydrocarbon receptor AICDA NM_020661.1 activation-induced cytidine deaminase AIRE NM_000383.2 autoimmune regulator APP NM_000484.3 amyloid beta (A4) precursor protein ARG1 NM_000045.2 arginase, liver ARG2 NM_001172.3 arginase, type II ARHGDIB NM_001175.4 Rho GDP dissociation inhibitor (GDI) beta ATG10 NM_001131028.1 ATG10 autophagy related 10 homolog (S. cerevisiae) ATG12 NM_004707.2 ATG12 autophagy related 12 homolog (S. cerevisiae) ATG16L1 NM_198890.2 ATG16 autophagy related 16-like 1 (S. cerevisiae) ATG5 NM_004849.2 ATG5 autophagy related 5 homolog (S. cerevisiae) ATG7 NM_001136031.2 ATG7 autophagy related 7 homolog (S. cerevisiae) ATM NM_000051.3 ataxia telangiectasia mutated B2M NM_004048.2 beta-2-microglobulin B3GAT1 NM_018644.3 beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) BATF NM_006399.3 basic leucine zipper transcription factor, ATF-like BATF3 NM_018664.2 basic leucine zipper transcription factor, ATF-like 3 BAX NM_138761.3 BCL2-associated X protein BCAP31 NM_005745.7 B-cell receptor-associated protein 31 BCL10 NM_003921.2 B-cell CLL/lymphoma 10 BCL2 NM_000657.2 B-cell CLL/lymphoma 2 BCL2L11 NM_138621.4 BCL2-like 11 (apoptosis facilitator) BCL3 NM_005178.2 B-cell CLL/lymphoma 3 BCL6 NM_001706.2 B-cell CLL/lymphoma 6 BID NM_001196.2 BH3 interacting domain death agonist BLNK NM_013314.2 -
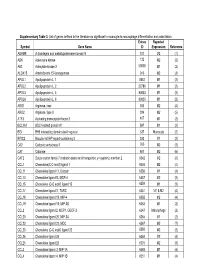
Supplementary Table 3. List of Genes Defined in the Literature As Significant in Monocyte-To-Macrophage Differentiation and Polarization
Supplementary Table 3. List of genes defined in the literature as significant in monocyte-to-macrophage differentiation and polarization. Entrez Reported Symbol Gene Name ID Expression Reference ADAM8 A disintegrin and metalloproteinase domain 8 101 M2 (1) ADK Adenosine kinase 132 M2 (2) AK3 Adenylate kinase 3 50808 M1 (2) ALOX15 Arachidonate 15-lipoxygenase 246 M2 (3) APOL1 Apolipoprotein L, 1 8542 M1 (2) APOL2 Apolipoprotein L, 2 23780 M1 (2) APOL3 Apolipoprotein L, 3 80833 M1 (2) APOL6 Apolipoprotein L, 6 80830 M1 (2) ARG1 Arginase, liver 383 M2 (4) ARG2 Arginase, type II 384 M2 (5) ATF3 Activating transcription factor 3 467 M1 (2) BCL2A1 BCL2-related protein A1 597 M1 (2) BID BH3 interacting domain death agonist 637 Monocyte (2) BIRC3 Baculoviral IAP repeat-containing 3 330 M1 (2) CA2 Carbonic anhydrase II 760 M2 (2) CAT Catalase 847 M2 (6) CAT2 Solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 6542 M2 (6) CCL1 Chemokine(C-C motif) ligand 1 6346 M2 (4) CCL11 Chemokine ligand 11, Eotaxin 6356 M1 (4) CCL13 Chemokine ligand13, MCP-4 6357 M2 (2) CCL15 Chemokine (C-C motif) ligand 15 6359 M1 (2) CCL17 Chemokine ligand17, TARC 6361 M1 & M2 (4) CCL18 Chemokine ligand 18, MIP-4 6362 M2 (4) CCL19 Chemokine ligand 19, MIP-3B 6363 M1 (2) CCL2 Chemokine ligand 2, MCP1, GDCF-2 6347 Macrophage (2) CCL20 Chemokine ligand 20, MIP-3A 6364 M1 (2) CCL22 Chemokine ligand 22, MDC 6367 M2 (7) CCL23 Chemokine (C-C motif) ligand 23 6368 M2 (2) CCL24 Chemokine ligand 24 6369 M2 (4) CCL25 Chemokine ligand 25 6370 M2 (8) CCL3 Chemokine