Generated by SRI International Pathway Tools Version 25.0, Authors S
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Generate Metabolic Map Poster
Authors: Pallavi Subhraveti Ron Caspi Peter Midford Peter D Karp An online version of this diagram is available at BioCyc.org. Biosynthetic pathways are positioned in the left of the cytoplasm, degradative pathways on the right, and reactions not assigned to any pathway are in the far right of the cytoplasm. Transporters and membrane proteins are shown on the membrane. Ingrid Keseler Periplasmic (where appropriate) and extracellular reactions and proteins may also be shown. Pathways are colored according to their cellular function. Gcf_001591825Cyc: Bacillus vietnamensis NBRC 101237 Cellular Overview Connections between pathways are omitted for legibility. Anamika Kothari sn-glycerol phosphate phosphate pro phosphate phosphate phosphate thiamine molybdate D-xylose D-ribose glutathione 3-phosphate D-mannitol L-cystine L-djenkolate lanthionine α,β-trehalose phosphate phosphate [+ 3 more] α,α-trehalose predicted predicted ABC ABC FliY ThiT XylF RbsB RS10935 UgpC TreP PutP RS10200 PstB PstB RS10385 RS03335 RS20030 RS19075 transporter transporter of molybdate of phosphate α,β-trehalose 6-phosphate L-cystine D-xylose D-ribose sn-glycerol D-mannitol phosphate phosphate thiamine glutathione α α phosphate phosphate phosphate phosphate L-djenkolate 3-phosphate , -trehalose 6-phosphate pro 1-phosphate lanthionine molybdate phosphate [+ 3 more] Metabolic Regulator Amino Acid Degradation Amine and Polyamine Biosynthesis Macromolecule Modification tRNA-uridine 2-thiolation Degradation ATP biosynthesis a mature peptidoglycan a nascent β an N-terminal- -

The Effect of Skin Fatty Acids on Staphylococcus Aureus
Arch Microbiol (2015) 197:245–267 DOI 10.1007/s00203-014-1048-1 ORGINAL PAPER The effect of skin fatty acids on Staphylococcus aureus Yvonne Neumann · Knut Ohlsen · Stefanie Donat · Susanne Engelmann · Harald Kusch · Dirk Albrecht · Michael Cartron · Alexander Hurd · Simon J. Foster Received: 23 July 2014 / Revised: 19 September 2014 / Accepted: 6 October 2014 / Published online: 18 October 2014 © The Author(s) 2014. This article is published with open access at Springerlink.com Abstract Staphylococcus aureus is a commensal of the hlgBC) was reduced, whereas that of host defence evasion human nose and skin. Human skin fatty acids, in particular components (cap, sspAB, katA) was increased. In particular, cis-6-hexadecenoic acid (C-6-H), have high antistaphylococ- members of the SaeRS regulon had highly reduced expres- cal activity and can inhibit virulence determinant production. sion, and the use of specific mutants revealed that the effect Here, we show that sub-MIC levels of C-6-H result in induc- on toxin production is likely mediated via SaeRS. tion of increased resistance. The mechanism(s) of C-6-H activity was investigated by combined transcriptome and Keywords S. aureus · Skin fatty acid · C-6-H · proteome analyses. Proteome analysis demonstrated a pleio- Resistance tropic effect of C-6-H on virulence determinant production. In response to C-6-H, transcriptomics revealed altered expres- sion of over 500 genes, involved in many aspects of virulence Introduction and cellular physiology. The expression of toxins (hla, hlb, The Gram-positive bacterium Staphylococcus aureus is able to survive as a commensal organism in the anterior nares and Communicated by Djamel DRIDER. -

ABSTRACT BOZDAG, AHMET. Investigation of Methanol and Formaldehyde Metabolism in Bacillus Methanolicus
ABSTRACT BOZDAG, AHMET. Investigation of Methanol and Formaldehyde Metabolism in Bacillus methanolicus.(Under the direction of Prof. Michael C. Flickinger). Bacillus methanolicus is a Gram-positive aerobic methylotroph growing optimally at 50-53 °C. Wild-type strains of B. methanolicus have been reported to secrete 58 g/l of L- glutamate in fed-batch cultures. Mutants of B. methanolicus created via classical mutangenesis can secrete 37 g/l of L-lysine, at 50 °C. The genes required for methylotrophyin B. methanolicus are encoded by an endogenous plasmid, pBM19 in strain MGA3, except for hexulose phosphate synthase (hps) and phosphohexuloisomerase (phi) which are encoded on the chromosome.It is a promising candidate for industrial production of chemical intermediates or amino acids from methanol. B. methanolicus employs the ribulose monophospate (RuMP) pathway to assimilate the carbon derived from the methanol, but enzymes that dissimilate carbon are not identified, although formaldehyde and formate were identified as intermediates by 13C NMR. It is important to understand how methanol is oxidized to formaldehyde and then, to formate and carbon dioxide. This study aims to elucidate the methanol dissimilation pathway of B. methanolicus. Growth rates of B. methanolicus MGA3 were assessed on methanol, mannitol, and glucose as a substrate. B. methanolicus achieved maximum growth rate, µmax, when growing on 25 mM methanol, 0.65±0.007 h-1, and it gradually decreased to 0.231±0.004 h-1 at 2Mmethanol concentration which demonstrates substrate inhibition. The maximum growth rates (µmax) of B. methanolicus MGA3 on mannitol and glucose are 0.532±0.002 and 0.336±0.003 h-1, respectively. -

12) United States Patent (10
US007635572B2 (12) UnitedO States Patent (10) Patent No.: US 7,635,572 B2 Zhou et al. (45) Date of Patent: Dec. 22, 2009 (54) METHODS FOR CONDUCTING ASSAYS FOR 5,506,121 A 4/1996 Skerra et al. ENZYME ACTIVITY ON PROTEIN 5,510,270 A 4/1996 Fodor et al. MICROARRAYS 5,512,492 A 4/1996 Herron et al. 5,516,635 A 5/1996 Ekins et al. (75) Inventors: Fang X. Zhou, New Haven, CT (US); 5,532,128 A 7/1996 Eggers Barry Schweitzer, Cheshire, CT (US) 5,538,897 A 7/1996 Yates, III et al. s s 5,541,070 A 7/1996 Kauvar (73) Assignee: Life Technologies Corporation, .. S.E. al Carlsbad, CA (US) 5,585,069 A 12/1996 Zanzucchi et al. 5,585,639 A 12/1996 Dorsel et al. (*) Notice: Subject to any disclaimer, the term of this 5,593,838 A 1/1997 Zanzucchi et al. patent is extended or adjusted under 35 5,605,662 A 2f1997 Heller et al. U.S.C. 154(b) by 0 days. 5,620,850 A 4/1997 Bamdad et al. 5,624,711 A 4/1997 Sundberg et al. (21) Appl. No.: 10/865,431 5,627,369 A 5/1997 Vestal et al. 5,629,213 A 5/1997 Kornguth et al. (22) Filed: Jun. 9, 2004 (Continued) (65) Prior Publication Data FOREIGN PATENT DOCUMENTS US 2005/O118665 A1 Jun. 2, 2005 EP 596421 10, 1993 EP 0619321 12/1994 (51) Int. Cl. EP O664452 7, 1995 CI2O 1/50 (2006.01) EP O818467 1, 1998 (52) U.S. -
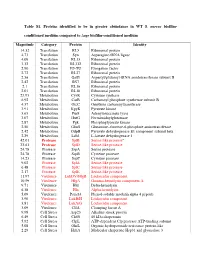
Conditioned Medium Compared to ∆Agr Biofilm-Conditioned Medium
Table S1. Proteins identified to be in greater abundance in WT S. aureus biofilm- conditioned medium compared to ∆agr biofilm-conditioned medium Magnitude Category Protein Identity 14.32 Translation RL5 Ribosomal protein 4.73 Translation Syn Asparagine-tRNA ligase 4.09 Translation RL13 Ribosomal protein 3.32 Translation RL332 Ribosomal protein 2.90 Translation EF-TU Elongation factor 2.72 Translation RL27 Ribosomal protein 2.56 Translation GatB Aspartyl/glutamyl-tRNA amidotransferase subunit B 2.42 Translation RS7 Ribosomal protein 2.1 Translation RL16 Ribosomal protein 2.01 Translation RL10 Ribosomal protein 25.93 Metabolism CysK Cysteine synthase 6.92 Metabolism CarB Carbamoyl phosphate synthetase subunit B 4.37 Metabolism OtcC Ornithine carbamoyltransferase 3.91 Metabolism KpyK Pyruvate kinase 3.63 Metabolism Pur8 Adenylosuccinate lyase 3.07 Metabolism HutG Formimidoylglutamase 2.87 Metabolism Pgk Phosphoglycerate kinase 2.50 Metabolism GlmS Glutamine--fructose-6-phosphate aminotransferase 2.42 Metabolism OdpB Pyruvate dehydrogenase E1 component subunit beta 2.29 Metabolism Ldh1 L-lactate dehydrogenase 1 42.01 Protease SplB Serine-like protease a 33.61 Protease SplD Serine-like protease 24.78 Protease SspA Serine protease 24.78 Protease SspB Cysteine protease 14.23 Protease SspP Cysteine protease 9.02 Protease SplA Serine-like protease 6.48 Protease SplC Serine-like protease 2.17 Protease SplE Serine-like protease 11.97 Virulence LukDV/HlgB Leukocidin component 10.59 Virulence HlgA Gamma-hemolysin component A 9.71 Virulence Hld Delta-hemolysin -

The Type VII Secretion System Protects Staphylococcus Aureus Against Antimicrobial Host Fatty Acids Arnaud Kengmo Tchoupa1, Kate E
www.nature.com/scientificreports OPEN The type VII secretion system protects Staphylococcus aureus against antimicrobial host fatty acids Arnaud Kengmo Tchoupa1, Kate E. Watkins1, Rebekah A. Jones1, Agnès Kuroki2, Mohammad Tauqeer Alam1, Sebastien Perrier1,2,3, Yin Chen4 & Meera Unnikrishnan1* The Staphylococcus aureus type VII secretion system (T7SS) exports several proteins that are pivotal for bacterial virulence. The mechanisms underlying T7SS-mediated staphylococcal survival during infection nevertheless remain unclear. Here we report that S. aureus lacking T7SS components are more susceptible to host-derived antimicrobial fatty acids. Unsaturated fatty acids such as linoleic acid (LA) elicited an increased inhibition of S. aureus mutants lacking T7SS efectors EsxC, EsxA and EsxB, or the membrane-bound ATPase EssC, compared to the wild-type (WT). T7SS mutants generated in diferent S. aureus strain backgrounds also displayed an increased sensitivity to LA. Analysis of bacterial membrane lipid profles revealed that the esxC mutant was less able to incorporate LA into its membrane phospholipids. Although the ability to bind labelled LA did not difer between the WT and mutant strains, LA induced more cell membrane damage in the T7SS mutants compared to the WT. Furthermore, proteomic analyses of WT and mutant cell fractions revealed that, in addition to compromising membranes, T7SS defects induce oxidative stress and hamper their response to LA challenge. Thus, our fndings indicate that T7SS contribute to maintaining S. aureus membrane integrity and homeostasis when bacteria encounter antimicrobial fatty acids. Staphylococcus aureus is a facultative pathogen that can colonize the skin and nares of healthy individuals. Te asymptomatic carriage of S. aureus is a major risk for subsequent infections 1. -
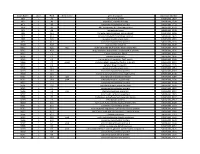
NTML 384-Well Array Well Identification
Strain Name plate Well Gene name gene discription Accession Number NE1 1 A1 cell surface protein SAUSA300_1327 NE2 1 A3 peptidase, rhomboid family SAUSA300_1509 NE3 1 A5 conserved hypothetical protein SAUSA300_1733 NE4 1 A7 ABC transporter ATP-binding protein SAUSA300_0309 NE5 1 A9 aminotransferase SAUSA300_2539 NE6 1 A11 formate dehydrogenase, alpha subunit SAUSA300_2258 NE7 1 A13 conserved hypothetical phage protein SAUSA300_1425 NE8 1 A15 putative membrane protein SAUSA300_1481 NE9 1 A17 conserved hypothetical protein SAUSA300_1294 NE10 1 A19 putative hemolysin III SAUSA300_2129 NE11 1 A21 recJ single-stranded-DNA-specific exonuclease RecJ SAUSA300_1592 NE12 1 A23 drug resistance transporter, EmrB/QacA subfamily SAUSA300_2126 NE13 1 C1 ribose transporter RbsU SAUSA300_0264 NE14 1 C3 putative transporter SAUSA300_2406 NE15 1 C5 transcriptional regulator, TetR family SAUSA300_2509 NE16 1 C7 moaD molybdopterin converting factor, subunit 1 SAUSA300_2221 NE17 1 C9 putative drug transporter SAUSA300_1705 NE18 1 C11 putative membrane protein SAUSA300_1851 NE19 1 C13 conserved hypothetical protein SAUSA300_0369 NE20 1 C15 transcriptional antiterminator, BglG family SAUSA300_0238 NE21 1 C17 upp uracil phosphoribosyltransferase SAUSA300_2066 NE22 1 C19 polA DNA polymerase I superfamily SAUSA300_1636 NE23 1 C21 conserved hypothetical protein SAUSA300_0740 NE24 1 C23 secretory antigen precursor SsaA SAUSA300_2503 NE25 1 E1 conserved hypothetical protein SAUSA300_1905 NE26 1 E3 coa staphylocoagulase precursor SAUSA300_0224 NE27 1 E5 nitric oxide -

1 the Type VII Secretion System Protects Staphylococcus Aureus Against
bioRxiv preprint doi: https://doi.org/10.1101/572172; this version posted February 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 The type VII secretion system protects Staphylococcus aureus against 2 antimicrobial host fatty acids 3 Arnaud Kengmo Tchoupa1, Kate E. Watkins1, Rebekah A. Jones1, Agnès Kuroki2, 4 Mohammad Tauqeer Alam1, Sebastien Perrier1,2,3, Yin Chen4, Meera Unnikrishnan1* 5 1Warwick Medical School, University of Warwick, Coventry, United Kingdom 6 2Department of Chemistry, University of Warwick, Coventry, United Kingdom 7 3Faculty of Pharmacy and Pharmaceutical Sciences, Monash University, Parkville, 8 Victoria 3052, Australia 9 4School of Life Sciences, University of Warwick, Coventry, United Kingdom 10 11 Running title: S. aureus T7SS and fatty acid toxicity 12 13 14 15 16 *Corresponding author 17 Email: [email protected]. 1 bioRxiv preprint doi: https://doi.org/10.1101/572172; this version posted February 16, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 18 Summary 19 The Staphylococcus aureus type VII secretion system (T7SS) exports several proteins 20 that are pivotal for bacterial virulence. The mechanisms underlying T7SS-mediated 21 staphylococcal survival during infection nevertheless remain unclear. Here we show 22 that the absence of EsxC, a small secreted effector implicated in bacterial persistence, 23 results in cell membrane defects in S. -

(12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur Et Al
US 2012026.6329A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0266329 A1 Mathur et al. (43) Pub. Date: Oct. 18, 2012 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/10 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/24 (2006.01) CI2N 9/02 (2006.01) (75) Inventors: Eric J. Mathur, Carlsbad, CA CI2N 9/06 (2006.01) (US); Cathy Chang, San Marcos, CI2P 2L/02 (2006.01) CA (US) CI2O I/04 (2006.01) CI2N 9/96 (2006.01) (73) Assignee: BP Corporation North America CI2N 5/82 (2006.01) Inc., Houston, TX (US) CI2N 15/53 (2006.01) CI2N IS/54 (2006.01) CI2N 15/57 2006.O1 (22) Filed: Feb. 20, 2012 CI2N IS/60 308: Related U.S. Application Data EN f :08: (62) Division of application No. 1 1/817,403, filed on May AOIH 5/00 (2006.01) 7, 2008, now Pat. No. 8,119,385, filed as application AOIH 5/10 (2006.01) No. PCT/US2006/007642 on Mar. 3, 2006. C07K I4/00 (2006.01) CI2N IS/II (2006.01) (60) Provisional application No. 60/658,984, filed on Mar. AOIH I/06 (2006.01) 4, 2005. CI2N 15/63 (2006.01) Publication Classification (52) U.S. Cl. ................... 800/293; 435/320.1; 435/252.3: 435/325; 435/254.11: 435/254.2:435/348; (51) Int. Cl. 435/419; 435/195; 435/196; 435/198: 435/233; CI2N 15/52 (2006.01) 435/201:435/232; 435/208; 435/227; 435/193; CI2N 15/85 (2006.01) 435/200; 435/189: 435/191: 435/69.1; 435/34; CI2N 5/86 (2006.01) 435/188:536/23.2; 435/468; 800/298; 800/320; CI2N 15/867 (2006.01) 800/317.2: 800/317.4: 800/320.3: 800/306; CI2N 5/864 (2006.01) 800/312 800/320.2: 800/317.3; 800/322; CI2N 5/8 (2006.01) 800/320.1; 530/350, 536/23.1: 800/278; 800/294 CI2N I/2 (2006.01) CI2N 5/10 (2006.01) (57) ABSTRACT CI2N L/15 (2006.01) CI2N I/19 (2006.01) The invention provides polypeptides, including enzymes, CI2N 9/14 (2006.01) structural proteins and binding proteins, polynucleotides CI2N 9/16 (2006.01) encoding these polypeptides, and methods of making and CI2N 9/20 (2006.01) using these polynucleotides and polypeptides. -

All Enzymes in BRENDA™ the Comprehensive Enzyme Information System
All enzymes in BRENDA™ The Comprehensive Enzyme Information System http://www.brenda-enzymes.org/index.php4?page=information/all_enzymes.php4 1.1.1.1 alcohol dehydrogenase 1.1.1.B1 D-arabitol-phosphate dehydrogenase 1.1.1.2 alcohol dehydrogenase (NADP+) 1.1.1.B3 (S)-specific secondary alcohol dehydrogenase 1.1.1.3 homoserine dehydrogenase 1.1.1.B4 (R)-specific secondary alcohol dehydrogenase 1.1.1.4 (R,R)-butanediol dehydrogenase 1.1.1.5 acetoin dehydrogenase 1.1.1.B5 NADP-retinol dehydrogenase 1.1.1.6 glycerol dehydrogenase 1.1.1.7 propanediol-phosphate dehydrogenase 1.1.1.8 glycerol-3-phosphate dehydrogenase (NAD+) 1.1.1.9 D-xylulose reductase 1.1.1.10 L-xylulose reductase 1.1.1.11 D-arabinitol 4-dehydrogenase 1.1.1.12 L-arabinitol 4-dehydrogenase 1.1.1.13 L-arabinitol 2-dehydrogenase 1.1.1.14 L-iditol 2-dehydrogenase 1.1.1.15 D-iditol 2-dehydrogenase 1.1.1.16 galactitol 2-dehydrogenase 1.1.1.17 mannitol-1-phosphate 5-dehydrogenase 1.1.1.18 inositol 2-dehydrogenase 1.1.1.19 glucuronate reductase 1.1.1.20 glucuronolactone reductase 1.1.1.21 aldehyde reductase 1.1.1.22 UDP-glucose 6-dehydrogenase 1.1.1.23 histidinol dehydrogenase 1.1.1.24 quinate dehydrogenase 1.1.1.25 shikimate dehydrogenase 1.1.1.26 glyoxylate reductase 1.1.1.27 L-lactate dehydrogenase 1.1.1.28 D-lactate dehydrogenase 1.1.1.29 glycerate dehydrogenase 1.1.1.30 3-hydroxybutyrate dehydrogenase 1.1.1.31 3-hydroxyisobutyrate dehydrogenase 1.1.1.32 mevaldate reductase 1.1.1.33 mevaldate reductase (NADPH) 1.1.1.34 hydroxymethylglutaryl-CoA reductase (NADPH) 1.1.1.35 3-hydroxyacyl-CoA -

IDENTIFICATION of NATIVE 20S PROTEASOMAL SUBSTRATES in the HALOARCHAEON Haloferax Volcanii THROUGH DEGRADOMIC and PHOSPHOPROTEOMIC ANALYSIS
IDENTIFICATION OF NATIVE 20S PROTEASOMAL SUBSTRATES IN THE HALOARCHAEON Haloferax volcanii THROUGH DEGRADOMIC AND PHOSPHOPROTEOMIC ANALYSIS By PHILLIP A. KIRKLAND A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2007 1 © 2007 Phillip Kirkland 2 To my mother, Kelly Kirkland; my grandmother Judy Jansen; and to the loving memory of Mr. and Mrs. L.V. Stevens. 3 ACKNOWLEDGMENTS I would like to express my deepest gratitude to Dr. Julie Maupin for her patience and guidance. Many thanks go to the Maupin lab staff (Matt Humbard, Wei Li, and Guanguin Zhou) and to former graduates Dr. Chris Reuter and Gosia Gil for all of their help and support. My sincerest thanks go to my supervisory committee (Dr. James Preston, Dr. Madeline Rasche, Dr. Richard Lamont, Dr. Peter Kima and Dr. Eric Triplett) for their insight. I owe a debt of gratitude to John Moore, Phi Do, Dr. Franz St. John, Youngnyun Kim, John Rice and the entire Microbiology and Cell Science Department. I am also extremely grateful to all of those who have offered help and support with instrumentation, bioinformatics and general proteomic endeavors including Dr. Stan Stevens, Scott McClung, Dr. Marjorie Chow and Dr. Alfred Chung and the entire ICBR staff as well as Dr. Jennifer Busby and Valerie Cavett at Scripps Florida. Finally, I am eternally grateful for the support and encouragement of my family and friends: my mom, Kelly Kirkland, my grandmother Judy Jansen, my fiancé Shelley Cooper, Jeremy and Matt Jones, William Knapp, and the Smith, Bright, Blair, Stevens and Jansen families and my great grandparents, the late Lavert V. -

(12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur Et Al
US 20150240226A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2015/0240226A1 Mathur et al. (43) Pub. Date: Aug. 27, 2015 (54) NUCLEICACIDS AND PROTEINS AND CI2N 9/16 (2006.01) METHODS FOR MAKING AND USING THEMI CI2N 9/02 (2006.01) CI2N 9/78 (2006.01) (71) Applicant: BP Corporation North America Inc., CI2N 9/12 (2006.01) Naperville, IL (US) CI2N 9/24 (2006.01) CI2O 1/02 (2006.01) (72) Inventors: Eric J. Mathur, San Diego, CA (US); CI2N 9/42 (2006.01) Cathy Chang, San Marcos, CA (US) (52) U.S. Cl. CPC. CI2N 9/88 (2013.01); C12O 1/02 (2013.01); (21) Appl. No.: 14/630,006 CI2O I/04 (2013.01): CI2N 9/80 (2013.01); CI2N 9/241.1 (2013.01); C12N 9/0065 (22) Filed: Feb. 24, 2015 (2013.01); C12N 9/2437 (2013.01); C12N 9/14 Related U.S. Application Data (2013.01); C12N 9/16 (2013.01); C12N 9/0061 (2013.01); C12N 9/78 (2013.01); C12N 9/0071 (62) Division of application No. 13/400,365, filed on Feb. (2013.01); C12N 9/1241 (2013.01): CI2N 20, 2012, now Pat. No. 8,962,800, which is a division 9/2482 (2013.01); C07K 2/00 (2013.01); C12Y of application No. 1 1/817,403, filed on May 7, 2008, 305/01004 (2013.01); C12Y 1 1 1/01016 now Pat. No. 8,119,385, filed as application No. PCT/ (2013.01); C12Y302/01004 (2013.01); C12Y US2006/007642 on Mar. 3, 2006.