Phenotypic and Phylogenetic Identification of Coliform Bacteria Obtained from 12 USEPA Approved Coliform Methods
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Get Consistent and Reliable Results
Need help selecting? M Microbiology Free Tech Support 800-323-4340 Cole-Parmer® Dehydrated Culture Media Get consistent and reliable results – Use when ready or dehydrated culture extends shelf life – Individually numbered with certificate of compliance expiration date, and shelf-life guarantee – Quick dissolve for faster processing – For laboratory use only Media, Dehydrated Media, Media Description Size (g) Catalog number Price Agar, bacteriological This clear gel agar is high grade and manufactured using the ice method 500 GH-14200-10 Azide dextrose broth Use for detection of fecal Streptococci in water and sewage 500 GH-14202-00 Baird parker agar Use for the selective isolation of coagulase positive Staphylococci in food 500 GH-14202-01 Blood agar base with low pH Use to cultivate a wide variety of bacteria 500 GH-14202-02 This is used as a differential medium in the isolation and presumptive identification of Bile Esculin agar 500 GH-14202-03 enterococci/group D streptococci Brain heart infusion agar Use to cultivate a wide spectrum of bacteria, yeasts, and molds 500 GH-14200-12 Brain heart infusion broth Use to cultivate fastidious aerobic and anaerobic bacteria 500 GH-14202-05 Brilliant green bile broth A standard methods medium for confirmed and completed tests for coliforms in water 500 GH-14200-14 Buffered peptone water Helps in the recovery of Salmonella from foods after preservation techniques 500 GH-14202-08 Cetrimide agar Use for the selective isolation of Pseudomonas aeruginosa 500 GH-14200-52 This broth is especially suited for environmental sampling where neutralization of the chemical D/E neutralizing Broth 500 GH-14202-11 is important to determining the bactericidal activity Use to isolate E. -

Prepared Culture Media
PREPARED CULTURE MEDIA 121517SS PREPARED CULTURE MEDIA Made in the USA AnaeroGRO™ DuoPak A 02 Bovine Blood Agar, 5%, with Esculin 13 AnaeroGRO™ DuoPak B 02 Bovine Blood Agar, 5%, with Esculin/ AnaeroGRO™ BBE Agar 03 MacConkey Biplate 13 AnaeroGRO™ BBE/PEA 03 Bovine Selective Strep Agar 13 AnaeroGRO™ Brucella Agar 03 Brucella Agar with 5% Sheep Blood, Hemin, AnaeroGRO™ Campylobacter and Vitamin K 13 Selective Agar 03 Brucella Broth with 15% Glycerol 13 AnaeroGRO™ CCFA 03 Brucella with H and K/LKV Biplate 14 AnaeroGRO™ Egg Yolk Agar, Modified 03 Buffered Peptone Water 14 AnaeroGRO™ LKV Agar 03 Buffered Peptone Water with 1% AnaeroGRO™ PEA 03 Tween® 20 14 AnaeroGRO™ MultiPak A 04 Buffered NaCl Peptone EP, USP 14 AnaeroGRO™ MultiPak B 04 Butterfield’s Phosphate Buffer 14 AnaeroGRO™ Chopped Meat Broth 05 Campy Cefex Agar, Modified 14 AnaeroGRO™ Chopped Meat Campy CVA Agar 14 Carbohydrate Broth 05 Campy FDA Agar 14 AnaeroGRO™ Chopped Meat Campy, Blood Free, Karmali Agar 14 Glucose Broth 05 Cetrimide Select Agar, USP 14 AnaeroGRO™ Thioglycollate with Hemin and CET/MAC/VJ Triplate 14 Vitamin K (H and K), without Indicator 05 CGB Agar for Cryptococcus 14 Anaerobic PEA 08 Chocolate Agar 15 Baird-Parker Agar 08 Chocolate/Martin Lewis with Barney Miller Medium 08 Lincomycin Biplate 15 BBE Agar 08 CompactDry™ SL 16 BBE Agar/PEA Agar 08 CompactDry™ LS 16 BBE/LKV Biplate 09 CompactDry™ TC 17 BCSA 09 CompactDry™ EC 17 BCYE Agar 09 CompactDry™ YMR 17 BCYE Selective Agar with CAV 09 CompactDry™ ETB 17 BCYE Selective Agar with CCVC 09 CompactDry™ YM 17 BCYE -

View Our Full Water Sampling Vials Product Offering
TABLE OF CONTENTS Environmental Monitoring 1 Sample Prep and Dilution 8 Dehydrated Culture Media - Criterion™ 12 Prepared Culture Media 14 Chromogenic Media - HardyCHROM™ 18 CompactDry™ 20 Quality Control 24 Membrane Filtration 25 Rapid Tests 26 Lab Supplies/Sample Collection 27 Made in the USA Headquarters Distribution Centers 1430 West McCoy Lane Santa Maria, California Santa Maria, CA 93455 Olympia, Washington 800.266.2222 : phone Salt Lake City, Utah [email protected] Phoenix, Arizona HardyDiagnostics.com Dallas, Texas Springboro, Ohio Hardy Diagnostics has a Quality Lake City, Florida Management System that is certified Albany, New York to ISO 13485 and is a FDA licensed Copyright © 2020 Hardy Diagnostics Raleigh, North Carolina medical device manufacturer. Environmental Monitoring Hardy Diagnostics offers a wide selection of quality products intended to help keep you up to date with regulatory compliance, ensure the efficacy of your cleaning protocol, and properly monitor your environment. 800.266.2222 [email protected] HardyDiagnostics.com 1 Environmental Monitoring Air Sampling TRIO.BAS™ Impact Air Samplers introduced the newest generation of microbial air sampling. These ergonomically designed instruments combine precise air sampling with modern connectivity to help you properly assess the air quality of your laboratory and simplify your process. MONO DUO Each kit includes: Each kit includes: TRIO.BAS™ MONO air sampler, induction TRIO.BAS™ DUO air sampler, battery battery charger and cable, aspirating charger -

BD Industry Catalog
PRODUCT CATALOG INDUSTRIAL MICROBIOLOGY BD Diagnostics Diagnostic Systems Table of Contents Table of Contents 1. Dehydrated Culture Media and Ingredients 5. Stains & Reagents 1.1 Dehydrated Culture Media and Ingredients .................................................................3 5.1 Gram Stains (Kits) ......................................................................................................75 1.1.1 Dehydrated Culture Media ......................................................................................... 3 5.2 Stains and Indicators ..................................................................................................75 5 1.1.2 Additives ...................................................................................................................31 5.3. Reagents and Enzymes ..............................................................................................75 1.2 Media and Ingredients ...............................................................................................34 1 6. Identification and Quality Control Products 1.2.1 Enrichments and Enzymes .........................................................................................34 6.1 BBL™ Crystal™ Identification Systems ..........................................................................79 1.2.2 Meat Peptones and Media ........................................................................................35 6.2 BBL™ Dryslide™ ..........................................................................................................80 -

150259-ID-None.Pdf
DIPONEGORO JOURNAL OF MAQUARES Volume 5, Nomor 3, Tahun 2016, Halaman: 157-164 MANAGEMENT OF AQUATIC RESOURCES http://ejournal-s1.undip.ac.id/index.php/maquares ANALISIS TOTAL BAKTERI COLIFORM DI PERAIRAN MUARA KALI WISO JEPARA The Analysis of Total Coliform Bacteria in Kali Wiso Estuary Jepara Wiwid Widyaningsih, Supriharyono *), Niniek Widyorini Program Studi Manajemen Sumberdaya Perairan Fakultas Perikanan dan Ilmu Kelautan, Universitas Diponegoro Jl. Prof. Soedarto, SH, Tembalang, Semarang, Jawa Tengah – 50275, Telp/Fax. +6224 7474698 Email : [email protected] ABSTRAK Kali Wiso merupakan sungai yang berada di tengah kota Jepara. Perairan ini menjadi tempat pembuangan limbah-limbah secara langsung. Limbah tersebut diantaranya limbah domestik, limbah pasar, limbah kapal, serta limbah TPI. Berdasarkan masukan limbah tersebut menjadikan muara ini tercemar. Perairan yang tercemar dapat dilihat dari pengamatan secara fisika, kimia, maupun biologis. Kondisi perairan yang tercemar secara biologis dilihat dari keberadaan bakteri patogen yang ada di perairan. Indikator bakteri yang digunakan yaitu bakteri coliform, karena sifatnya yang berkorelasi positif dengan bakteri patogen lainnya. Pemanfaatan perairan ini digunakan untuk kegiatan pelabuhan, tempat bersandar kapal nelayan, serta kegiatan perikanan yang ada di sekitar perairan Jepara. Oleh karena itu perlu diketahui kepadatan bakteri coliform sehingga dapat bermanfaat sesuai dengan peruntukannya. Tujuan dari penelitian ini adalah untuk mengetahui total bakteri coliform serta mengetahui adanya bakteri Escherichia coli. Penelitain ini dilakukan pada bulan Maret 2016 di Muara Kali Wiso dengan dua kali pengulangan dalam kondisi pasang dan surut. Metode yang digunakan yaitu survei dengan teknik sampling purposive sampling. Metode analisa laboratorium yang digunakan berdasarkan SNI -01-2332- 1991. Kepadatan bakteri coliform pada perairan muara Kali Wiso yaitu >110.000 sel/100ml dan bakteri Escherichia coli sebesar >110.000 sel/100ml. -
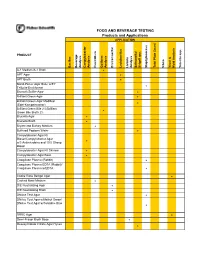
DONE Food and Beverage Testing
FOOD AND BEVERAGE TESTING Products and Applications APPLICATION PRODUCT Bacillus Beverage Analysis Campylobacter Analysis Clostridium Coliform Analysis Environmental Lactobacillus Listeria Analysis Salmonella/ Shigell spp. Staphylococcus Total Plate Count Vibrio Yeast & Mold Analysis Yersinia spp. A-1 Medium/A-1 Broth ! APT Agar ! APT Broth ! Baird-Parker Agar Base w/EY Tellurite Enrichment ! Bismuth Sulfite Agar ! Brilliant Green Agar ! Brilliant Green Agar Modified (Edel-Kampelmacher) ! Brilliant Green Bile 2%/Brilliant Green Bile Broth 2% ! Brucella Agar ! Brucella Broth ! Bryant and Burkey Medium ! Buffered Peptone Water ! Campylobacter Agar Kit Blaser/Campylobacter Agar w/5 Antimicrobics and 10% Sheep ! Blood Campylobacter Agar Kit Skirrow ! Campylobacter Agar Base ! Coagulase Plasma (Rabbit) ! Coagulase Plasma EDTA (Rabbit)/ Coagulase Plasma w/EDTA ! Cooke Rose Bengal Agar ! Cooked Meat Medium ! D/E Neutralizing Agar ! D/E Neutralizing Broth ! DNAse Test Agar ! DNAse Test Agar w/Methyl Green/ DNAse Test Agar w/Toluidine Blue ! DRBC Agar ! Demi-Fraser Broth Base ! Desoxycholate Citrate Agar Hynes ! FOOD AND BEVERAGE TESTING Products and Applications APPLICATION PRODUCT Bacillus Beverage Analysis Campylobacter Analysis Clostridium Coliform Analysis Environmental Lactobacillus Listeria Analysis Salmonella/ Shigell spp. Staphylococcus Total Plate Count Vibrio Yeast & Mold Analysis Yersinia spp. Differential Reinforced Clostridial Agar ! EC Medium/EC Broth ! EC Medium with MUG/EC Broth w/MUG ! Elliker Broth ! m Endo Agar LES ! m Endo -

Method 1680: Fecal Coliforms in Sewage Sludge (Biosolids) by Multiple-Tube Fermentation Using Lauryl Tryptose Broth (LTB) and EC Medium
United States Office of Water EPA-821-R-14-009 Environmental Protection Agency (4303-T) September 2014 Method 1680: Fecal Coliforms in Sewage Sludge (Biosolids) by Multiple-Tube Fermentation using Lauryl Tryptose Broth (LTB) and EC Medium U.S. Environmental Protection Agency Office of Water (4303T) 1200 Pennsylvania Avenue, NW Washington, DC 20460 Method 1680 Acknowledgments The contributions of the following persons and organizations to this study are gratefully acknowledged: Referee Laboratory • EPA Office of Research and Development, National Risk Management Research Lab: Mark C. Meckes and Karen M. White Volunteer Participant Laboratories • American Interplex: John Overbey and Lizbeth Huggins • BioVir Laboratories: Rick Danielson and Jim Truscott • City of Los Angeles Bureau of Sanitation Environmental Monitoring Division: Farhana Mohamed and Zora Bahariance • County Sanitation Districts of Los Angeles County: Shawn Thompson and Julie Millenbach • Environmental Associates: Susan Boutros and John Chandler • Hampton Roads Sanitation District: Anna Rule and Bob Maunz • King County Environmental Laboratory: Greg Ma and Bobbie Anderson • Hoosier Microbiological Laboratories: Don Hendrickson, Keri Nixon, Katy Bilger, and Lindsey Shelton • Massachusetts Water Resources Authority: Steve Rhode and Mariya Gofshteyn • Milwaukee Metropolitan Sewerage District: Jeff MacDonald and Tim O’Neill • University of Iowa Hygienic Laboratory: Nancy Hall and Cathy Lord • Utah Department of Health: Sanwat Chaudhuri and Devon Cole The following facilities provided -

Protective Effects of Probiotics on Cognitive and Motor Functions, Anxiety Level, Visceral Sensitivity, Oxidative Stress And
life Article Protective Effects of Probiotics on Cognitive and Motor Functions, Anxiety Level, Visceral Sensitivity, Oxidative Stress and Microbiota in Mice with Antibiotic-Induced Dysbiosis Alisa Arslanova 1, Aksiniya Tarasova 1, Anastasia Alexandrova 2, Vera Novoselova 2, Ilnar Shaidullov 1 , Dilyara Khusnutdinova 3, Tatiana Grigoryeva 3, Dina Yarullina 2, Olga Yakovleva 1 and Guzel Sitdikova 1,* 1 Department of Human and Animal Physiology, Institute of Fundamental Medicine and Biology, Kazan Federal University, 420008 Kazan, Republic of Tatarstan, Russia; [email protected] (A.A.); [email protected] (A.T.); [email protected] (I.S.); [email protected] (O.Y.) 2 Department of Microbiology, Institute of Fundamental Medicine and Biology, Kazan Federal University, 420008 Kazan, Republic of Tatarstan, Russia; [email protected] (A.A.); [email protected] (V.N.); [email protected] (D.Y.) 3 “Omics Technologies” Laboratory, Institute of Fundamental Medicine and Biology, Kazan Federal University, 420008 Kazan, Republic of Tatarstan, Russia; [email protected] (D.K.); [email protected] (T.G.) * Correspondence: [email protected] Citation: Arslanova, A.; Tarasova, A.; Abstract: Accumulating clinical and preclinical data indicate a prominent role of gut microbiota in Alexandrova, A.; Novoselova, V.; regulation of physiological functions. The gut-brain axis imbalance due to gut dysbiosis is associated Shaidullov, I.; Khusnutdinova, D.; with a range of neurodegenerative diseases. Probiotics were suggested not only to restore intestinal Grigoryeva, T.; Yarullina, D.; dysbiosis but also modulate stress response and improve mood and anxiety symptoms. In this study, Yakovleva, O.; Sitdikova, G. we assessed the effects of probiotic lactobacilli on behavioral reactions, the level of oxidative stress Protective Effects of Probiotics on and microbiota content in mice administered to broad-spectrum antibiotics. -

Differences in the Composition of Cultivable Aerobic and Facultative Anaerobic Oral Microbiota in Cats of Various Age Groups
DOI: 10.2478/fv-2021-0009 FOLIA VETERINARIA, 65, 1: 67—74, 2021 DIFFERENCES IN THE COMPOSITION OF CULTIVABLE AEROBIC AND FACULTATIVE ANAEROBIC ORAL MICROBIOTA IN CATS OF VARIOUS AGE GROUPS Sondorová, M., Koščová, J., Kačírová, J., Maďar, M. Department of Microbiology and Immunology, University of Veterinary Medicine and Pharmacy in Košice, Komenského 73, 041 81 Košice Slovakia [email protected] ABSTRACT oral micro biota were examined, differences between age groups were noted. The microbial diversity of the oral The feline oral cavity is naturally inhabited by var microbiota significantly increased with age. ious microorganisms contributing to the maintenance of its oral health. The imbalance of oral microbiota or Key words: age groups; cultivation methods; feline; the presence of pathogenic agents can lead to secondary oral microbiota oral diseases. Various factors such as sex, diet, breed, environment and even age, affect the composition of a healthy oral microbiota during the life of cats. The INTRODUCTION purpose of this study was to compare the composition of culturable aerobic and facultative anaerobic micro The oral cavity is the first part of the gastrointestinal biota in cats in terms of different age categories. We tract where the process of digestion begins, and therefore used conventional cultivation methods in conjunction it creates a space for the action of various microorganisms with microscopic and biochemical methods to isolate [8]. The constant flow of saliva and the unique biological and identify the micro organisms found in the oral cavi properties of each part of the oral cavity provide a place for ty of cats. The examination of 76 samples confirmed the attachment of microorganisms [22]. -

International Catalogue 2014
Microbiology Microbiology IVD IVD INTERNATIONALINTERNATIONAL CATACATALOGUELOGUE 2014 Microbiology In Vitro Diagnostics Viale Monza 272, 20128 Milan, Italy Biolife Leader in Microbiology 2 1 20162 0 6 CHROMOGENICCHHROOMOG6 CRE 1992 19929 C-EC AGARAGA [ \ 2 1 20152 0 5 of E.coli 5 CHROMOGENICCHHROOMOG ESBL for the detection of ESBL 1996 19961 1968 ALOA 19 19869 86 [ Dehydrated culture 6 medium for the media manufacturing. MUCAPMUCCAP TESTTE detection and [ enumeration of Hektoen Enteric Agar, L.monocytogenes Giolitti & Cantoni Broth, for Salmonella, made Staphylococci 110 Medium: Biolife 2 1 20132 0 3 [ 3 Biolife. CHROMOGENICCHHROOMOG STREP B for the detection of S.agalactiae 19 2 19 19821 1985 8 85 SELECTIVE MICROBIOLOGY8 MUG MEDIA 199898 19989 Biolife [ CHROMOGENICOMOGENIC Biolife SALMONELLA AGAR (CSA) [ culture media for the detection of 2 12 20122 0 Salmonella 0 CHROMOGENICCHHROOMOG CANDIDA AGAR for the detection of clinically important Candida! 2 2 20002 2006 000 006 IMMUNOCROMATHOGRAPHICIM0MUNOCR TESTS SENECA [ the enumeration of E.coli + Enterobacteriaceae Some of our milestones INTERNATIONAL CATALOGUE PRODUCTS FOR MICROBIOLOGY IN VITRO DIAGNOSTICS Biolife Italiana S.r.l. Mascia Brunelli S.p.A. International Catalogue - Finito di stampare nel mese di ottobre 2017 presso INGRAF srl (MI) - Rev. 11/2017 Biolife - Mascia Brunelli International Catalogue Biolife Italiana S.r.l. and Mascia Brunelli Spa are two sisters companies belonging to the same Group, operating, since long time, in the fields of Microbiology, Immunological IVDs and Medical Devices. Biolife Italiana S.r.l. has a well-established reputation in the production of powdered and ready-to use culture media for microbiology, supplements and enrichments, kits and reagents for microbial identifi- cation. -

Mississippi Academy of Sciences
MISSISSIPPI ACADEMY OF SCIENCES EIGHTY-FOURTH ANNUAL MEETING February 20-21, 2020 Mississippi Gulf Coast and Convention Center Biloxi, MS Sponsors (Annual Meeting Sponsor) Mississippi State University- College of Agriculture and Life Sciences College of Forest Resources James Worth Bagley College of Engieering Journal of the Mississippi Academy of Sciences Volume 65 January 2020 Number 1 Contents 3 ACADEMY OFFICERS & DIVISION CHAIRS 2019-2020 Editor 4 GENERAL SCHEDULE Michelle Tucci University of Mississippi Medical Center 7 DIRECTIONS TO CONFERENCE CENTER Editorial Board 8 SUSTAINING MEMBERS 9 LIFE MEMBERS Maria Begonia Jackson State University 10 EXHIBITORS 11 EXECUTIVE DIRECTOR’S COLUMN Ibrahim O. Farah 12 MAS AWARDS AND DODGEN LECTURE Jackson State University 16 SPECIAL PRESENTATIONS Millsaps Undergraduate Student Awards and Mississippi INBRE Robin Rockhold Graduate Student Awards, LSMAMP University of Mississippi Medical Center Ham Benghuzzi 24 OVERVIEW OF DIVISIONAL PROGRAMS University of Mississippi Medical Center Biloxi Convention Center Floor Plans and Notes Program Editor ABSTRACTS Kenneth Butler 36 Agriculture and Plant Science University of Mississippi Medical Center 43 Cellular, Molecular and Developmental Biology 63 Chemistry and Chemical Engineering The Journal of the Mississippi Academy of Sciences (ISSN 0076-9436) is published in 88 Ecology and Evolutionary Biology January (annual meeting abstracts), April, 94 Geology and Geography July, and October, by the Mississippi Acad- emy of Sciences. Members of the Academy 101 Health Sciences receive the journal as part of their regular (nonstudent) membership. Inquiries 123 History and Philosophy of Science regarding subscriptions, availability of back 127 Marine and Atmospheric Sciences issues, and address changes should be addressed to The Mississippi Academy of 130 Mathematics, Computer Science and Statistics Sciences, Post Office Box 55709, Jackson, 136 Physics and Engineering MS 39296-5709, telephone 601-977-0627, or email [email protected]. -

CDC Anaerobe 5% Sheep Blood Agar with Phenylethyl Alcohol (PEA) CDC Anaerobe Laked Sheep Blood Agar with Kanamycin and Vancomycin (KV)
Difco & BBL Manual Manual of Microbiological Culture Media Second Edition Editors Mary Jo Zimbro, B.S., MT (ASCP) David A. Power, Ph.D. Sharon M. Miller, B.S., MT (ASCP) George E. Wilson, MBA, B.S., MT (ASCP) Julie A. Johnson, B.A. BD Diagnostics – Diagnostic Systems 7 Loveton Circle Sparks, MD 21152 Difco Manual Preface.ind 1 3/16/09 3:02:34 PM Table of Contents Contents Preface ...............................................................................................................................................................v About This Manual ...........................................................................................................................................vii History of BD Diagnostics .................................................................................................................................ix Section I: Monographs .......................................................................................................................................1 History of Microbiology and Culture Media ...................................................................................................3 Microorganism Growth Requirements .............................................................................................................4 Functional Types of Culture Media ..................................................................................................................5 Culture Media Ingredients – Agars ...................................................................................................................6