The Existing Literature in Connection with the Bacteriology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

NCTC) Bacterial Strain Equivalents to American Type Culture Collection (ATCC) Bacterial Strains
This list shows National Collection of Type Cultures (NCTC) bacterial strain equivalents to American Type Culture Collection (ATCC) bacterial strains. NCTC Number CurrentName ATCC Number NCTC 7212 Acetobacter pasteurianus ATCC 23761 NCTC 10138 Acholeplasma axanthum ATCC 25176 NCTC 10171 Acholeplasma equifetale ATCC 29724 NCTC 10128 Acholeplasma granularum ATCC 19168 NCTC 10172 Acholeplasma hippikon ATCC 29725 NCTC 10116 Acholeplasma laidlawii ATCC 23206 NCTC 10134 Acholeplasma modicum ATCC 29102 NCTC 10188 Acholeplasma morum ATCC 33211 NCTC 10150 Acholeplasma oculi ATCC 27350 NCTC 10198 Acholeplasma parvum ATCC 29892 NCTC 8582 Achromobacter denitrificans ATCC 15173 NCTC 10309 Achromobacter metalcaligenes ATCC 17910 NCTC 10807 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 27061 NCTC 10808 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 17062 NCTC 10809 Achromobacter xylosoxidans subsp. xylosoxidans ATCC 27063 NCTC 12156 Acinetobacter baumannii ATCC 19606 NCTC 10303 Acinetobacter baumannii ATCC 17904 NCTC 7844 Acinetobacter calcoaceticus ATCC 15308 NCTC 12983 Acinetobacter calcoaceticus ATCC 23055 NCTC 8102 acinetobacter dna group 13 ATCC 17903 NCTC 10304 Acinetobacter genospecies 13 ATCC 17905 NCTC 10306 Acinetobacter haemolyticus ATCC 17907 NCTC 10305 Acinetobacter haemolyticus subsp haemolyticus ATCC 17906 NCTC 10308 Acinetobacter johnsonii ATCC 17909 NCTC 10307 Acinetobacter junii ATCC 17908 NCTC 5866 Acinetobacter lwoffii ATCC 15309 NCTC 12870 Actinobacillus delphinicola ATCC 700179 NCTC 8529 Actinobacillus equuli ATCC 19392 -

Legionella Gresilensis Sp. Nov. and Legionella Beliardensis Sp. Nov., Isolated from Water in France
International Journal of Systematic and Evolutionary Microbiology (2001), 51, 1949–1957 Printed in Great Britain Legionella gresilensis sp. nov. and Legionella beliardensis sp. nov., isolated from water in France 1 Centre National de Franc: ois Lo Presti,1‡ Serge Riffard,1 He! le' ne Meugnier,1 Re! fe! rence des Legionella 1 2 3 UPRES EA1655, Faculte! de Monique Reyrolle, Yves Lasne, † Patrick A. D. Grimont, Me! decine RTH Laennec, Francine Grimont,3 Robert F. Benson,4 Don J. Brenner,4 Rue Guillaume Paradin, 4 1 1 69372 Lyon cedex 08, Arnold G. Steigerwalt, Jerome Etienne and Jean Freney France 2 Laboratoire des Author for correspondence: Franc: ois Lo Presti. Tel: j33 0 169 79 79 60. Fax: j33 0 169 79 79 20. Radioisotopes et de e-mail: Francois.Lo-Presti!sanofi-synthelabo.com Biochimie mole! culaire, Ho# pital Edouard Herriot, ! ! 69437 Lyon cedex 03, Novel Legionella-like isolates, strains Montbeliard A1T and Greoux 11 D13T, France isolated from two different French water sources, were studied taxonomically 3 Unite! des Ente! robacte! ries, and phylogenetically. Morphological and biochemical characterization revealed Institut Pasteur, 75724 Paris cedex 15, France that they were Gram-negative, aerobic, non-spore-forming bacilli with a cut- glass appearance that grew only on L-cysteine-supplemented buffered charcoal 4 Respiratory Diseases Branch, Meningitis and yeast extract agar. Phenotypic characterization using fatty acid and ubiquinone Special Pathogens Branch, profiles and SDS-PAGE analysis confirmed that they were closely related, but National Center for distinct from, other species of the genus Legionella, since serotyping could not Infectious Diseases, Centers for Disease Control and relate them to any existing serogroup. -

Which Organisms Are Used for Anti-Biofouling Studies
Table S1. Semi-systematic review raw data answering: Which organisms are used for anti-biofouling studies? Antifoulant Method Organism(s) Model Bacteria Type of Biofilm Source (Y if mentioned) Detection Method composite membranes E. coli ATCC25922 Y LIVE/DEAD baclight [1] stain S. aureus ATCC255923 composite membranes E. coli ATCC25922 Y colony counting [2] S. aureus RSKK 1009 graphene oxide Saccharomycetes colony counting [3] methyl p-hydroxybenzoate L. monocytogenes [4] potassium sorbate P. putida Y. enterocolitica A. hydrophila composite membranes E. coli Y FESEM [5] (unspecified/unique sample type) S. aureus (unspecified/unique sample type) K. pneumonia ATCC13883 P. aeruginosa BAA-1744 composite membranes E. coli Y SEM [6] (unspecified/unique sample type) S. aureus (unspecified/unique sample type) graphene oxide E. coli ATCC25922 Y colony counting [7] S. aureus ATCC9144 P. aeruginosa ATCCPAO1 composite membranes E. coli Y measuring flux [8] (unspecified/unique sample type) graphene oxide E. coli Y colony counting [9] (unspecified/unique SEM sample type) LIVE/DEAD baclight S. aureus stain (unspecified/unique sample type) modified membrane P. aeruginosa P60 Y DAPI [10] Bacillus sp. G-84 LIVE/DEAD baclight stain bacteriophages E. coli (K12) Y measuring flux [11] ATCC11303-B4 quorum quenching P. aeruginosa KCTC LIVE/DEAD baclight [12] 2513 stain modified membrane E. coli colony counting [13] (unspecified/unique colony counting sample type) measuring flux S. aureus (unspecified/unique sample type) modified membrane E. coli BW26437 Y measuring flux [14] graphene oxide Klebsiella colony counting [15] (unspecified/unique sample type) P. aeruginosa (unspecified/unique sample type) graphene oxide P. aeruginosa measuring flux [16] (unspecified/unique sample type) composite membranes E. -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -
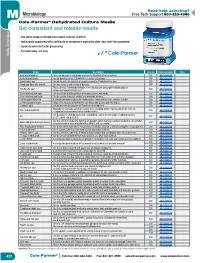
Get Consistent and Reliable Results
Need help selecting? M Microbiology Free Tech Support 800-323-4340 Cole-Parmer® Dehydrated Culture Media Get consistent and reliable results – Use when ready or dehydrated culture extends shelf life – Individually numbered with certificate of compliance expiration date, and shelf-life guarantee – Quick dissolve for faster processing – For laboratory use only Media, Dehydrated Media, Media Description Size (g) Catalog number Price Agar, bacteriological This clear gel agar is high grade and manufactured using the ice method 500 GH-14200-10 Azide dextrose broth Use for detection of fecal Streptococci in water and sewage 500 GH-14202-00 Baird parker agar Use for the selective isolation of coagulase positive Staphylococci in food 500 GH-14202-01 Blood agar base with low pH Use to cultivate a wide variety of bacteria 500 GH-14202-02 This is used as a differential medium in the isolation and presumptive identification of Bile Esculin agar 500 GH-14202-03 enterococci/group D streptococci Brain heart infusion agar Use to cultivate a wide spectrum of bacteria, yeasts, and molds 500 GH-14200-12 Brain heart infusion broth Use to cultivate fastidious aerobic and anaerobic bacteria 500 GH-14202-05 Brilliant green bile broth A standard methods medium for confirmed and completed tests for coliforms in water 500 GH-14200-14 Buffered peptone water Helps in the recovery of Salmonella from foods after preservation techniques 500 GH-14202-08 Cetrimide agar Use for the selective isolation of Pseudomonas aeruginosa 500 GH-14200-52 This broth is especially suited for environmental sampling where neutralization of the chemical D/E neutralizing Broth 500 GH-14202-11 is important to determining the bactericidal activity Use to isolate E. -

Prepared Culture Media
PREPARED CULTURE MEDIA 121517SS PREPARED CULTURE MEDIA Made in the USA AnaeroGRO™ DuoPak A 02 Bovine Blood Agar, 5%, with Esculin 13 AnaeroGRO™ DuoPak B 02 Bovine Blood Agar, 5%, with Esculin/ AnaeroGRO™ BBE Agar 03 MacConkey Biplate 13 AnaeroGRO™ BBE/PEA 03 Bovine Selective Strep Agar 13 AnaeroGRO™ Brucella Agar 03 Brucella Agar with 5% Sheep Blood, Hemin, AnaeroGRO™ Campylobacter and Vitamin K 13 Selective Agar 03 Brucella Broth with 15% Glycerol 13 AnaeroGRO™ CCFA 03 Brucella with H and K/LKV Biplate 14 AnaeroGRO™ Egg Yolk Agar, Modified 03 Buffered Peptone Water 14 AnaeroGRO™ LKV Agar 03 Buffered Peptone Water with 1% AnaeroGRO™ PEA 03 Tween® 20 14 AnaeroGRO™ MultiPak A 04 Buffered NaCl Peptone EP, USP 14 AnaeroGRO™ MultiPak B 04 Butterfield’s Phosphate Buffer 14 AnaeroGRO™ Chopped Meat Broth 05 Campy Cefex Agar, Modified 14 AnaeroGRO™ Chopped Meat Campy CVA Agar 14 Carbohydrate Broth 05 Campy FDA Agar 14 AnaeroGRO™ Chopped Meat Campy, Blood Free, Karmali Agar 14 Glucose Broth 05 Cetrimide Select Agar, USP 14 AnaeroGRO™ Thioglycollate with Hemin and CET/MAC/VJ Triplate 14 Vitamin K (H and K), without Indicator 05 CGB Agar for Cryptococcus 14 Anaerobic PEA 08 Chocolate Agar 15 Baird-Parker Agar 08 Chocolate/Martin Lewis with Barney Miller Medium 08 Lincomycin Biplate 15 BBE Agar 08 CompactDry™ SL 16 BBE Agar/PEA Agar 08 CompactDry™ LS 16 BBE/LKV Biplate 09 CompactDry™ TC 17 BCSA 09 CompactDry™ EC 17 BCYE Agar 09 CompactDry™ YMR 17 BCYE Selective Agar with CAV 09 CompactDry™ ETB 17 BCYE Selective Agar with CCVC 09 CompactDry™ YM 17 BCYE -

View Our Full Water Sampling Vials Product Offering
TABLE OF CONTENTS Environmental Monitoring 1 Sample Prep and Dilution 8 Dehydrated Culture Media - Criterion™ 12 Prepared Culture Media 14 Chromogenic Media - HardyCHROM™ 18 CompactDry™ 20 Quality Control 24 Membrane Filtration 25 Rapid Tests 26 Lab Supplies/Sample Collection 27 Made in the USA Headquarters Distribution Centers 1430 West McCoy Lane Santa Maria, California Santa Maria, CA 93455 Olympia, Washington 800.266.2222 : phone Salt Lake City, Utah [email protected] Phoenix, Arizona HardyDiagnostics.com Dallas, Texas Springboro, Ohio Hardy Diagnostics has a Quality Lake City, Florida Management System that is certified Albany, New York to ISO 13485 and is a FDA licensed Copyright © 2020 Hardy Diagnostics Raleigh, North Carolina medical device manufacturer. Environmental Monitoring Hardy Diagnostics offers a wide selection of quality products intended to help keep you up to date with regulatory compliance, ensure the efficacy of your cleaning protocol, and properly monitor your environment. 800.266.2222 [email protected] HardyDiagnostics.com 1 Environmental Monitoring Air Sampling TRIO.BAS™ Impact Air Samplers introduced the newest generation of microbial air sampling. These ergonomically designed instruments combine precise air sampling with modern connectivity to help you properly assess the air quality of your laboratory and simplify your process. MONO DUO Each kit includes: Each kit includes: TRIO.BAS™ MONO air sampler, induction TRIO.BAS™ DUO air sampler, battery battery charger and cable, aspirating charger -

Aquascreen® Legionella Species Qpcr Detection Kit
AquaScreen® Legionella species qPCR Detection Kit INSTRUCTIONS FOR USE FOR USE IN RESEARCH AND QUALITY CONTROL Symbols Lot No. Cat. No. Expiry date Storage temperature Number of reactions Manufacturer INDICATION The AquaScreen® Legionella species qPCR Detection kit is specifically designed for the quantitative detection of several Legionella species in water samples prepared with the AquaScreen® FastExt- ract kit. Its design complies with the requirements of AFNOR T90-471 and ISO/TS 12869:2012. Legionella are ubiquitous bacteria in surface water and moist soil, where they parasitize protozoa. The optimal growth temperature lies between +15 and +45 °C, whereas these gram-negative bacteria are dormant below 20 °C and do not survive above 60 °C. Importantly, Legionella are well-known as opportunistic intracellular human pathogens causing Legionnaires’ disease and Pontiac fever. The transmission occurs through inhalation of contami- nated aerosols generated by an infected source (e.g. human-made water systems like shower- heads, sink faucets, heaters, cooling towers, and many more). In order to efficiently prevent Legionella outbreaks, water safety control measures need syste- matic application but also reliable validation by fast Legionella testing. TEST PRINCIPLE The AquaScreen® Legionella species Kit uses qPCR for quantitative detection of legionella in wa- ter samples. In contrast to more time-consuming culture-based methods, AquaScreen® assays need less than six hours including sample preparation and qPCR to reliably detect Legionella. Moreover, the AquaScreen® qPCR assay has proven excellent performance in terms of specificity and sensitivity: other bacterial genera remain undetected whereas linear quantification is obtai- ned up to 1 x 106 particles per sample, therefore requiring no material dilution. -

Immunoproteomic Identification of Biomarkers for Diagnosis of Legionellosis
Immunoproteomic identification of biomarkers for diagnosis of legionellosis Submitted in total fulfilment of the requirements for the degree of Doctor of philosophy by Kaylass Poorun Department of Chemistry and Biotechnology Faculty of Science, Engineering and Technology Swinburne University of Technology Australia 2014 Abstract Abstract Legionellosis, a disease with significant mortality and morbidity rates, is considered to be the second most frequent cause of severe community-acquired pneumonia. It is difficult to distinguish from other types of pneumonia due to similar clinical manifestations. Several studies have demonstrated the inadequacies of current diagnostic tests for confirming Legionella infections. This study was aimed at identifying biomarkers that can be used in an improved test. A comparative proteomic analysis, using DIGE, was carried out between L. pneumophila ATCC33152 and L. longbeachae NSW150 and D4968 isolates. While many homologous proteins were found to be commonly expressed, numerous others were identified to be differentially expressed under similar in vitro conditions suggesting that the two species have different lifestyles and infection strategies. The bacterial immunoglobulin domain containing protein, found to share sequence homology to Type V secretion proteins intimin and invasin, is not known to be present in Legionella. Human sera containing antibodies against Legionella from a set of blind samples were identified by ELISA. Downstream analyses revealed that diverse immunogens may be responsible for eliciting immune response in different Legionella species which in turn show little to no congeneric cross-reactivity. To the best of our knowledge, this is a unique finding not previously reported. Several serological diagnostic tests currently in use do not include many Legionella species in their testing panel, which may be a reason for many Legionella species being under-reported. -

BD Industry Catalog
PRODUCT CATALOG INDUSTRIAL MICROBIOLOGY BD Diagnostics Diagnostic Systems Table of Contents Table of Contents 1. Dehydrated Culture Media and Ingredients 5. Stains & Reagents 1.1 Dehydrated Culture Media and Ingredients .................................................................3 5.1 Gram Stains (Kits) ......................................................................................................75 1.1.1 Dehydrated Culture Media ......................................................................................... 3 5.2 Stains and Indicators ..................................................................................................75 5 1.1.2 Additives ...................................................................................................................31 5.3. Reagents and Enzymes ..............................................................................................75 1.2 Media and Ingredients ...............................................................................................34 1 6. Identification and Quality Control Products 1.2.1 Enrichments and Enzymes .........................................................................................34 6.1 BBL™ Crystal™ Identification Systems ..........................................................................79 1.2.2 Meat Peptones and Media ........................................................................................35 6.2 BBL™ Dryslide™ ..........................................................................................................80 -

Mitigating Biofouling on Reverse Osmosis Membranes Via Greener Preservatives
Mitigating biofouling on reverse osmosis membranes via greener preservatives by Anna Curtin Biology (BSc), Le Moyne College, 2017 A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of MASTER OF APPLIED SCIENCE in the Department of Civil Engineering, University of Victoria © Anna Curtin, 2020 University of Victoria All rights reserved. This Thesis may not be reproduced in whole or in part, by photocopy or other means, without the permission of the author. Supervisory Committee Mitigating biofouling on reverse osmosis membranes via greener preservatives by Anna Curtin Biology (BSc), Le Moyne College, 2017 Supervisory Committee Heather Buckley, Department of Civil Engineering Supervisor Caetano Dorea, Department of Civil Engineering, Civil Engineering Departmental Member ii Abstract Water scarcity is an issue faced across the globe that is only expected to worsen in the coming years. We are therefore in need of methods for treating non-traditional sources of water. One promising method is desalination of brackish and seawater via reverse osmosis (RO). RO, however, is limited by biofouling, which is the buildup of organisms at the water-membrane interface. Biofouling causes the RO membrane to clog over time, which increases the energy requirement of the system. Eventually, the RO membrane must be treated, which tends to damage the membrane, reducing its lifespan. Additionally, antifoulant chemicals have the potential to create antimicrobial resistance, especially if they remain undegraded in the concentrate water. Finally, the hazard of chemicals used to treat biofouling must be acknowledged because although unlikely, smaller molecules run the risk of passing through the membrane and negatively impacting humans and the environment. -

150259-ID-None.Pdf
DIPONEGORO JOURNAL OF MAQUARES Volume 5, Nomor 3, Tahun 2016, Halaman: 157-164 MANAGEMENT OF AQUATIC RESOURCES http://ejournal-s1.undip.ac.id/index.php/maquares ANALISIS TOTAL BAKTERI COLIFORM DI PERAIRAN MUARA KALI WISO JEPARA The Analysis of Total Coliform Bacteria in Kali Wiso Estuary Jepara Wiwid Widyaningsih, Supriharyono *), Niniek Widyorini Program Studi Manajemen Sumberdaya Perairan Fakultas Perikanan dan Ilmu Kelautan, Universitas Diponegoro Jl. Prof. Soedarto, SH, Tembalang, Semarang, Jawa Tengah – 50275, Telp/Fax. +6224 7474698 Email : [email protected] ABSTRAK Kali Wiso merupakan sungai yang berada di tengah kota Jepara. Perairan ini menjadi tempat pembuangan limbah-limbah secara langsung. Limbah tersebut diantaranya limbah domestik, limbah pasar, limbah kapal, serta limbah TPI. Berdasarkan masukan limbah tersebut menjadikan muara ini tercemar. Perairan yang tercemar dapat dilihat dari pengamatan secara fisika, kimia, maupun biologis. Kondisi perairan yang tercemar secara biologis dilihat dari keberadaan bakteri patogen yang ada di perairan. Indikator bakteri yang digunakan yaitu bakteri coliform, karena sifatnya yang berkorelasi positif dengan bakteri patogen lainnya. Pemanfaatan perairan ini digunakan untuk kegiatan pelabuhan, tempat bersandar kapal nelayan, serta kegiatan perikanan yang ada di sekitar perairan Jepara. Oleh karena itu perlu diketahui kepadatan bakteri coliform sehingga dapat bermanfaat sesuai dengan peruntukannya. Tujuan dari penelitian ini adalah untuk mengetahui total bakteri coliform serta mengetahui adanya bakteri Escherichia coli. Penelitain ini dilakukan pada bulan Maret 2016 di Muara Kali Wiso dengan dua kali pengulangan dalam kondisi pasang dan surut. Metode yang digunakan yaitu survei dengan teknik sampling purposive sampling. Metode analisa laboratorium yang digunakan berdasarkan SNI -01-2332- 1991. Kepadatan bakteri coliform pada perairan muara Kali Wiso yaitu >110.000 sel/100ml dan bakteri Escherichia coli sebesar >110.000 sel/100ml.