Endogenous GPCR Transcript Profiling of Life Technologies' Primary Cell Offerings: Opening the Door to Physiologically Relevan
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Missense Polymorphism in the Putative Pheromone Receptor Gene VN1R1 Is Associated with Sociosexual Behavior
OPEN Citation: Transl Psychiatry (2017) 7, e1102; doi:10.1038/tp.2017.70 www.nature.com/tp ORIGINAL ARTICLE A missense polymorphism in the putative pheromone receptor gene VN1R1 is associated with sociosexual behavior S Henningsson1, D Hovey1, K Vass1, H Walum2,3,4,5, K Sandnabba6, P Santtila6, P Jern6 and L Westberg1 Pheromones regulate social and reproductive behavior in most mammalian species. These effects are mediated by the vomeronasal and main olfactory systems. Effects of putative pheromones on human neuroendocrine activity, brain activity and attractiveness ratings suggest that humans may communicate via similar chemosignaling. Here we studied two samples of younger and older individuals, respectively, with respect to one nonsynonymous polymorphism in the gene encoding the human vomeronasal type-1 receptor 1, VN1R1, and one nonsynonymous polymorphism in the gene encoding the olfactory receptor OR7D4. Participants in both samples had self-reported their sociosexual behavior using the sociosexual orientation inventory, including questions regarding lifetime number of one-night stands, number of partners last year and expected number of partners the coming 5 years. In women, there was a significant association between the VN1R1 polymorphism and sociosexual behavior in both samples, driven specifically by the question regarding one-night stands. Our results support the hypothesis that human social interaction is modulated by communication via chemosignaling. Translational Psychiatry (2017) 7, e1102; doi:10.1038/tp.2017.70; published -

Download Download
Supplementary Figure S1. Results of flow cytometry analysis, performed to estimate CD34 positivity, after immunomagnetic separation in two different experiments. As monoclonal antibody for labeling the sample, the fluorescein isothiocyanate (FITC)- conjugated mouse anti-human CD34 MoAb (Mylteni) was used. Briefly, cell samples were incubated in the presence of the indicated MoAbs, at the proper dilution, in PBS containing 5% FCS and 1% Fc receptor (FcR) blocking reagent (Miltenyi) for 30 min at 4 C. Cells were then washed twice, resuspended with PBS and analyzed by a Coulter Epics XL (Coulter Electronics Inc., Hialeah, FL, USA) flow cytometer. only use Non-commercial 1 Supplementary Table S1. Complete list of the datasets used in this study and their sources. GEO Total samples Geo selected GEO accession of used Platform Reference series in series samples samples GSM142565 GSM142566 GSM142567 GSM142568 GSE6146 HG-U133A 14 8 - GSM142569 GSM142571 GSM142572 GSM142574 GSM51391 GSM51392 GSE2666 HG-U133A 36 4 1 GSM51393 GSM51394 only GSM321583 GSE12803 HG-U133A 20 3 GSM321584 2 GSM321585 use Promyelocytes_1 Promyelocytes_2 Promyelocytes_3 Promyelocytes_4 HG-U133A 8 8 3 GSE64282 Promyelocytes_5 Promyelocytes_6 Promyelocytes_7 Promyelocytes_8 Non-commercial 2 Supplementary Table S2. Chromosomal regions up-regulated in CD34+ samples as identified by the LAP procedure with the two-class statistics coded in the PREDA R package and an FDR threshold of 0.5. Functional enrichment analysis has been performed using DAVID (http://david.abcc.ncifcrf.gov/) -

Genetics of Aggressive Behavior: an Overview Kim Veroude,1 Yanli Zhang-James,2,3 No�Elia Fern�Andez-Castillo,4,5,6 Mireille J
RESEARCH ARTICLE Neuropsychiatric Genetics Genetics of Aggressive Behavior: An Overview Kim Veroude,1 Yanli Zhang-James,2,3 Noelia Fernandez-Castillo,4,5,6 Mireille J. Bakker,1 Bru Cormand,4,5,6 and Stephen V. Faraone2,3,7* 1Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition and Behaviour, Radboudumc, Nijmegen, The Netherlands 2Departments of Psychiatry and of Neuroscience and Physiology, SUNY Upstate Medical University, Syracuse, New York 3Departments of Neuroscience and Physiology, SUNY Upstate Medical University, Syracuse, New York 4Departament de Genetica, Facultat de Biologia, Universitat de Barcelona, Catalonia, Spain 5Institut de Biomedicina de la Universitat de Barcelona (IBUB), Catalonia, Spain 6Centro de Investigacion Biomedica en Red de Enfermedades Raras (CIBERER), Spain 7K.G. Jebsen Centre for Research on Neuropsychiatric Disorders, University of Bergen, Bergen, Norway Manuscript Received: 22 April 2015; Manuscript Accepted: 5 August 2015 The Research Domain Criteria (RDoC) address three types of aggression: frustrative non-reward, defensive aggression and How to Cite this Article: offensive/proactive aggression. This review sought to present Veroude K, Zhang-James Y, Fernandez- the evidence for genetic underpinnings of aggression and to Castillo N, Bakker MJ, Cormand B, determine to what degree prior studies have examined pheno- Faraone SV. 2016. Genetics of Aggressive types that fit into the RDoC framework. Although the constructs Behavior: An Overview. of defensive and offensive aggression have been widely used in the animal genetics literature, the human literature is mostly Am J Med Genet Part B 171B:3–43. agnostic with regard to all the RDoC constructs. We know from twin studies that about half the variance in behavior may be explained by genetic risk factors. -

A Web-Platform for Analysis of Host Factors Involved in Viral Infections Discovered by Genome Wide Rnai Screen
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is © The Royal Society of Chemistry 2017 vhfRNAi: A web-platform for analysis of host factors involved in viral infections discovered by genome wide RNAi screen Anamika Thakur#, Abid Qureshi# and Manoj Kumar* Bioinformatics Centre, Institute of Microbial Technology, Council of Scientific and Industrial Research, Sector 39A, Chandigarh-160036, India #Equal contribution * To whom correspondence should be addressed. Tel, 91-172-6665453; Fax, 91-172- 2690585; 91-172-2690632; Email, [email protected] Supplementary Tables Table S1: Statistics of unique and duplicate host factors in each virus Table S2: Table denoting genes common among different viruses Table S3: Statistics of GWAS analysis Table S1. Statistics of unique and duplicate host factors in each virus S. No. Virus Unique-Entries Duplicate-Entries 1. Adeno-associated virus (AAV) 926 533 2. Avian influenza virus (AIV) 0 11 3. Borna disease virus (BDV) 14 20 4. Dengue virus 2 (DEN-2) 27 13 5. Hepatitis C virus (HCV) 236 213 6. Human immunodeficiency virus 1 (HIV) 1388 857 7. Human parainfluenza virus 3 (HPIV-3) 0 27 8. Human herpesvirus 1 (HSV-1) 34 38 9. Influenza A virus (IAV) 700 513 10. Lymphocytic choriomeningitis virus (LCMV) 0 54 11. Marburgvirus (MARV) 0 11 12. Poliovirus (PV) 3340 1035 13. Rotavirus (RV) 347 175 14. Sendai virus (SeV) 32 27 15. Sindbis virus (SIV) 70 41 16. Vaccinia virus (VACV) 482 296 17. Vesicular stomatitis virus (VSV) 9 78 18. West Nile virus (WNV) 313 137 Table S1. Statistics of unique host factors for each virus having overlapping and unique factors in different viruses Non-overlap – Overall- S. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Genetics of Aggressive Behavior: an Overview
The influence of genes on “positive valence systems” constructs: A systematic review Item Type Article Authors Hess, Jonathan L.; Kawaguchi, Daniel M.; Wagner, Kayla E.; Faraone, Stephen V.; Glatt, Stephen J. Citation Hess, JL, Kawaguchi, DM, Wagner, KE, Faraone, SV, Glatt, SJ. 2015. The Influence of Genes on “Positive Valence Systems” Constructs: A Systematic Review. Am J Med Genet Part B 171B: 92– 110. DOI 10.1002/ajmg.b.32382 Publisher Wiley Rights Attribution-NonCommercial-NoDerivatives 4.0 International Download date 26/09/2021 01:12:21 Item License http://doi.wiley.com/10.1002/tdm_license_1.1 Link to Item http://hdl.handle.net/20.500.12648/1795 See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/281608624 Genetics of Aggressive Behavior: An Overview Article in American Journal of Medical Genetics Part B Neuropsychiatric Genetics · September 2015 DOI: 10.1002/ajmg.b.32364 CITATIONS READS 81 1,596 6 authors, including: Yanli Zhang-James Noèlia Fernàndez-Castillo State University of New York Upstate Medical University University of Barcelona 65 PUBLICATIONS 1,992 CITATIONS 102 PUBLICATIONS 848 CITATIONS SEE PROFILE SEE PROFILE Mireille J. Bakker Bru Cormand Radboud University Medical Centre (Radboudumc) University of Barcelona 8 PUBLICATIONS 272 CITATIONS 311 PUBLICATIONS 10,197 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Aggression Genetics View project Trastorno por deficit de atencion con hiperactividad en la clinica pediatrica View project All content following this page was uploaded by Noèlia Fernàndez-Castillo on 25 November 2017. The user has requested enhancement of the downloaded file. -

Structure and Function Analyses of the Purified GPCR Human Vomeronasal Type 1 Receptor 1
Structure and function analyses of the purified GPCR human vomeronasal type 1 receptor 1 The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation Corin, Karolina, Philipp Baaske, Sandra Geissler, Christoph J. Wienken, Stefan Duhr, Dieter Braun, and Shuguang Zhang. “Structure and Function Analyses of the Purified GPCR Human Vomeronasal Type 1 Receptor 1.” Sci. Rep. 1 (November 28, 2011). As Published http://dx.doi.org/10.1038/srep00172 Publisher Nature Publishing Group Version Final published version Citable link http://hdl.handle.net/1721.1/88274 Terms of Use Creative Commons Attribution-Noncommercial-Share Alike Detailed Terms http://creativecommons.org/licenses/by-nc-sa/3.0/ Structure and function analyses of the purified GPCR human vomeronasal SUBJECT AREAS: BIOCHEMISTRY type 1 receptor 1 MEMBRANE DYNAMICS Karolina Corin1, Philipp Baaske2, Sandra Geissler2, Christoph J. Wienken3, Stefan Duhr2, Dieter Braun3 MOLECULAR ENGINEERING & Shuguang Zhang1 BIOPHYSICS 1Center for Biomedical Engineering, NE47-379, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA Received 02139-4307, USA, 2NanoTemper Technologies GmbH, Amalienstrasse 54, 80799 Mu¨nchen, Germany, 3Systems Biophysics, 3 May 2011 Functional Nanosystems, Department of Physics, Ludwig-Maximilians University Mu¨nchen, Amalienstrasse 54, 80799 Mu¨nchen, Germany. Accepted 25 October 2011 Published The vomeronasal system is one of several fine-tuned scent-detecting signaling systems in mammals. However, despite significant efforts, how these receptors detect scent remains an enigma. One reason is the 28 November 2011 lack of sufficient purified receptors to perform detailed biochemical, biophysical and structural analyses. Here we report the ability to express and purify milligrams of purified, functional human vomeronasal receptor hVN1R1. -

Figure S1. RNA Degradation Map of the Three Gene Chips. Each of the 76 Samples Is Presented by a Differently Colored Line. Table SI
Figure S1. RNA degradation map of the three gene chips. Each of the 76 samples is presented by a differently colored line. Table SI. Upregulated (n=1,300) and downregulated (n=912) differentially expressed genes. A, Upregulated genes Gene Log fold Average t P-value Adjusted change expression P-value LINGO1 2.6777 6.4170 23.1699 5.54x10-37 1.69x10-33 VWF 1.6093 6.2905 23.0801 7.24x10-37 1.74x10-33 PYCR1 1.4290 7.4945 22.2201 9.80x10-36 1.93x10-32 DTL 2.7697 6.1038 21.9223 2.46x10-35 4.09x10-32 CKAP2L 1.9408 6.7911 21.8106 3.48x10-35 5.31x10-32 CASC5 1.0329 4.9592 21.7923 3.68x10-35 5.31x10-32 RDM1 1.8612 5.8981 21.4909 9.46x10-35 1.28x10-31 HMGA1 1.9524 7.0245 21.2584 1.97x10-34 2.37x10-31 GRHL2 2.3061 6.6285 21.2028 2.35x10-34 2.68x10-31 EMC3-AS1 1.7596 6.4341 20.7583 9.76x10-34 1.06x10-30 DSN1 1.5764 5.6077 20.7126 1.13x10-33 1.16x10-30 CENPF 2.0079 6.1039 20.5981 1.64x10-33 1.48x10-30 BIK 2.8614 6.9592 20.4699 2.48x10-33 2.15x10-30 CDCA8 1.8125 7.2037 20.3946 3.18x10-33 2.45x10-30 CCNB2 1.3002 6.2043 19.9845 1.22x10-32 8.81x10-30 DUSP9 1.7127 6.3549 19.8314 2.03x10-32 1.42x10-29 ESPL1 2.0699 7.4012 19.4946 6.27x10-32 4.11x10-29 CSPG5 2.0283 6.6777 19.3062 1.18x10-31 7.32x10-29 CDCA5 2.5265 7.5517 19.1695 1.88x10-31 1.10x10-28 KIF20B 2.2678 6.4109 19.0425 2.90x10-31 1.57x10-28 E2F1 1.4217 7.0710 19.0116 3.23x10-31 1.70x10-28 CDCA3 2.2727 7.4162 18.9327 4.23x10-31 1.99x10-28 MUC1 2.2947 7.7898 18.8818 5.04x10-31 2.27x10-28 KIF2C 1.6191 7.1943 18.7275 8.57x10-31 3.71x10-28 BIRC5 1.6937 6.5189 18.7158 8.92x10-31 3.78x10-28 CLPB 1.3245 6.0626 -

Program in Human Neutrophils Fails To
Downloaded from http://www.jimmunol.org/ by guest on September 25, 2021 is online at: average * The Journal of Immunology Anaplasma phagocytophilum , 20 of which you can access for free at: 2005; 174:6364-6372; ; from submission to initial decision 4 weeks from acceptance to publication J Immunol doi: 10.4049/jimmunol.174.10.6364 http://www.jimmunol.org/content/174/10/6364 Insights into Pathogen Immune Evasion Mechanisms: Fails to Induce an Apoptosis Differentiation Program in Human Neutrophils Dori L. Borjesson, Scott D. Kobayashi, Adeline R. Whitney, Jovanka M. Voyich, Cynthia M. Argue and Frank R. DeLeo cites 28 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2005/05/03/174.10.6364.DC1 This article http://www.jimmunol.org/content/174/10/6364.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* • Why • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2005 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 25, 2021. The Journal of Immunology Insights into Pathogen Immune Evasion Mechanisms: Anaplasma phagocytophilum Fails to Induce an Apoptosis Differentiation Program in Human Neutrophils1 Dori L. -

Genome-Wide Identification of Directed Gene Networks Using Large-Scale Population Genomics Data
ARTICLE DOI: 10.1038/s41467-018-05452-6 OPEN Genome-wide identification of directed gene networks using large-scale population genomics data René Luijk1, Koen F. Dekkers1, Maarten van Iterson1, Wibowo Arindrarto 2, Annique Claringbould3, Paul Hop1, BIOS Consortium#, Dorret I. Boomsma4, Cornelia M. van Duijn5, Marleen M.J. van Greevenbroek6,7, Jan H. Veldink8, Cisca Wijmenga3, Lude Franke 3, Peter A.C. ’t Hoen 9, Rick Jansen 10, Joyce van Meurs11, Hailiang Mei2, P. Eline Slagboom1, Bastiaan T. Heijmans 1 & Erik W. van Zwet12 1234567890():,; Identification of causal drivers behind regulatory gene networks is crucial in understanding gene function. Here, we develop a method for the large-scale inference of gene–gene interactions in observational population genomics data that are both directed (using local genetic instruments as causal anchors, akin to Mendelian Randomization) and specific (by controlling for linkage disequilibrium and pleiotropy). Analysis of genotype and whole-blood RNA-sequencing data from 3072 individuals identified 49 genes as drivers of downstream transcriptional changes (Wald P <7×10−10), among which transcription factors were over- represented (Fisher’s P = 3.3 × 10−7). Our analysis suggests new gene functions and targets, including for SENP7 (zinc-finger genes involved in retroviral repression) and BCL2A1 (target genes possibly involved in auditory dysfunction). Our work highlights the utility of population genomics data in deriving directed gene expression networks. A resource of trans-effects for all 6600 genes with a genetic instrument can be explored individually using a web-based browser. 1 Molecular Epidemiology Section, Department of Medical Statistics and Bioinformatics, Leiden University Medical Center, Leiden, Zuid-Holland 2333 ZC, The Netherlands. -
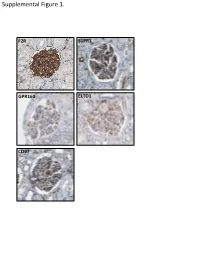
Supplemental Data
Supplemental Figure 1. F2R S1PR1 GPR160 ELTD1 CD97 Supplemental Figure 2. A B brain heart lung liver kidney spleen testis skm Expression in mouse ssues glom rok Gprc5a Gapdh Human Protein Atlas RNAseq database Supplemental Figure 3. A Gprc5a Pdgfrb Merged CL CL Gprc5a CD31 Merged CL CL B C 1.4 * 1.2 mc 1 matrix 0.8 0.6 0.4 matrix 0.2 0 pod end 1 2 mes3 D Vector Gprc5a E 40 kD - - Gprc5a - acn Supplemental Figure 4. Control 12 month-old KO 12 month-old A B C D E F 1 2 0.8 * 1.5 score 0.6 m µ 1 0.4 0.2 Slits/ 0.5 Mesangial 0 0 Ctrl KO Ctrl KO Supplemental Figure 5. A B 1.2 Vector Gprc5a 1 * 0.8 0.6 * 0.4 * Normalized Density 0.2 0 pEGFR/tEGR pSmad/tSmad TGF-β1 C 1.8 siCON 1.6 siGprc5a * 1.4 * * 1.2 1 0.8 0.6 NormalizedDensity 0.4 0.2 0 pEGFR/tEGR pSmad/tSmad TGF-β1 Supplemental table 1. List of glomerulus-expressed GPCRs as detected by qPCR. Data shown as mean ± standard deviation (Glom=glomerulus, Rok=rest of kidney). GPCR Glom Rok Glom/Rok LPAR6 41892,11 ± 38478,89 1040,06 ± 1370,12 39,28 ELTD1 30275,64 ± 14085,26 33,23 ± 46,99 910,13 GPR116 24020,06 ± 7789,84 55,1 ± 47,75 434,96 PTH1R 15402,81 ± 17644,32 7521,01 ± 3264,57 1,05 CALCRL 14096,09 ± 3854,84 199,06 ± 222,52 69,81 HPRT1 13342,04 ± 10677,69 1824,77 ± 1767,23 6,31 S1PR5 9474,7 ± 10124,3 110,84 ± 29,54 84,48 LPHN2 8645,89 ± 914,74 256,04 ± 293,87 32,77 FZD1 8176,2 ± 4947,45 1321,97 ± 1311,91 5,18 CXCR4 7097,31 ± 4388,91 535,98 ± 640,28 12,24 GPR160 6446,59 ± 1550,07 4816,3 ± 5918,99 0,34 NPY1R 6177,74 ± 6282,76 209,64 ± 296,48 28,47 PTGER4 5323,66 ± 3789,51 179,13 ± 210 28,72 RXFP1 -

Supplemental Material Supplemental Material and Methods
Supplemental Material Supplemental Material and Methods Library preparation and sequencing 24 FFPE samples suitable for library preparation according to quality/quantity evaluations were processed following the manufacturer’s specifications with minor modifications by using the Illumina TruSeq® RNA Access Library Prep (Illumina) along with purification steps employing SPRI beads. DV200 values for all samples exceeded 40%, and 100 ng of total RNA was used for the cDNA synthesis. Each library was quantified with the fluorimeter Qubit 2.0 (dsDNA HS kit; Thermo Fisher), and the size distribution was determined using a DNA 1000 kit on a 2100 Bioanalyzer instrument prior to pooling. All libraries had a similar size distribution of approximately 260 bp. A 4-plex pool of libraries was made by combining 200 ng of each DNA library. The libraries were sequenced on an Illumina NextSeq 500. At least 180 M pair-end 2x75 bp reads were generated per library. The libraries were prepared and sequenced at EMBL Genomics Core Facility (Heidelberg, Germany). NGS data analysis. The raw FASTQ sequencing files were aligned to human reference genome (build h19) with bowtie2 (version 2.3.2) and tophat2 (version 2.1). Differentially expressed genes (DEGs) between groups were determined using LIMA-R package with default settings. The MCP-counter R package was used to estimate the abundance of tissue-infiltrating immune cell populations. Supplemental figures Suppl. Fig. 1. Experimental design of the study. Suppl. Fig. 2 Representative images of CALR immunostaining. Scale bar = 100 μm. Suppl. Fig. 3. Flow cytometry-assisted quantification of surface exposed CALR. (A) Gating strategy. The percentage of cells in each gate is reported.