Figure S1. RNA Degradation Map of the Three Gene Chips. Each of the 76 Samples Is Presented by a Differently Colored Line. Table SI
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Missense Polymorphism in the Putative Pheromone Receptor Gene VN1R1 Is Associated with Sociosexual Behavior
OPEN Citation: Transl Psychiatry (2017) 7, e1102; doi:10.1038/tp.2017.70 www.nature.com/tp ORIGINAL ARTICLE A missense polymorphism in the putative pheromone receptor gene VN1R1 is associated with sociosexual behavior S Henningsson1, D Hovey1, K Vass1, H Walum2,3,4,5, K Sandnabba6, P Santtila6, P Jern6 and L Westberg1 Pheromones regulate social and reproductive behavior in most mammalian species. These effects are mediated by the vomeronasal and main olfactory systems. Effects of putative pheromones on human neuroendocrine activity, brain activity and attractiveness ratings suggest that humans may communicate via similar chemosignaling. Here we studied two samples of younger and older individuals, respectively, with respect to one nonsynonymous polymorphism in the gene encoding the human vomeronasal type-1 receptor 1, VN1R1, and one nonsynonymous polymorphism in the gene encoding the olfactory receptor OR7D4. Participants in both samples had self-reported their sociosexual behavior using the sociosexual orientation inventory, including questions regarding lifetime number of one-night stands, number of partners last year and expected number of partners the coming 5 years. In women, there was a significant association between the VN1R1 polymorphism and sociosexual behavior in both samples, driven specifically by the question regarding one-night stands. Our results support the hypothesis that human social interaction is modulated by communication via chemosignaling. Translational Psychiatry (2017) 7, e1102; doi:10.1038/tp.2017.70; published -
Critical Evaluation of Gene Expression Changes in Human Tissues In
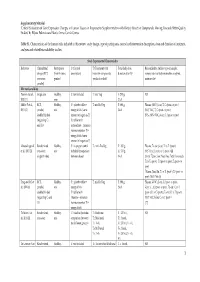
Supplementary Material ‘Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies’ by Biljana Pokimica and María-Teresa García-Conesa Table S1. Characteristics of the human trials included in this review: study design, type of participants, control and intervention description, dose and duration of treatment, analyses and related bioavailability studies. Study Experimental Characteristics Reference Clinical trial Participants C (Control T (Treatment with Total daily dose, Bioavailability studies: type of sample, design (RCT, (health status, description) bioactive compounds, duration (d or h)1 compounds and (or) metabolites analysed, crossover, gender) products or diet) main results2 parallel) Mix meals and diets Persson I et al., Single arm Healthy, C: not included T: mix Veg T: 250 g, NR 2000 [1] men 21 d Møller P et al., RCT, Healthy, C1: placebo tablet + T: mix FruVeg T: 600 g, Plasma: (NS↑) β-car, T, C2 (post- vs pre-) 2003 [2] parallel, mix energy drink (same 24 d (NC) VitC, T, C2 (post- vs pre-) double blinded amount of sugars as T) (NS↓, 69%) VitC, β-car, C1 (post- vs pre-) (regarding C1 C2: tablet with and C2) antioxidants + minerals (same amount as T) + energy drink (same amount of sugars as T) Almendingen K Randomized, Healthy, C: no proper control T1,2: mix FruVeg T1: 300 g, Plasma: ↑α-car, β-car, T2 vs T1 (post-) et al., 2005 [3] crossover, mix included (comparison T2: 750 g, (NS↑) Lyc, Lut, T2 vs T1 (post-) [4] single -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Download Download
Supplementary Figure S1. Results of flow cytometry analysis, performed to estimate CD34 positivity, after immunomagnetic separation in two different experiments. As monoclonal antibody for labeling the sample, the fluorescein isothiocyanate (FITC)- conjugated mouse anti-human CD34 MoAb (Mylteni) was used. Briefly, cell samples were incubated in the presence of the indicated MoAbs, at the proper dilution, in PBS containing 5% FCS and 1% Fc receptor (FcR) blocking reagent (Miltenyi) for 30 min at 4 C. Cells were then washed twice, resuspended with PBS and analyzed by a Coulter Epics XL (Coulter Electronics Inc., Hialeah, FL, USA) flow cytometer. only use Non-commercial 1 Supplementary Table S1. Complete list of the datasets used in this study and their sources. GEO Total samples Geo selected GEO accession of used Platform Reference series in series samples samples GSM142565 GSM142566 GSM142567 GSM142568 GSE6146 HG-U133A 14 8 - GSM142569 GSM142571 GSM142572 GSM142574 GSM51391 GSM51392 GSE2666 HG-U133A 36 4 1 GSM51393 GSM51394 only GSM321583 GSE12803 HG-U133A 20 3 GSM321584 2 GSM321585 use Promyelocytes_1 Promyelocytes_2 Promyelocytes_3 Promyelocytes_4 HG-U133A 8 8 3 GSE64282 Promyelocytes_5 Promyelocytes_6 Promyelocytes_7 Promyelocytes_8 Non-commercial 2 Supplementary Table S2. Chromosomal regions up-regulated in CD34+ samples as identified by the LAP procedure with the two-class statistics coded in the PREDA R package and an FDR threshold of 0.5. Functional enrichment analysis has been performed using DAVID (http://david.abcc.ncifcrf.gov/) -

Genetics of Aggressive Behavior: an Overview Kim Veroude,1 Yanli Zhang-James,2,3 No�Elia Fern�Andez-Castillo,4,5,6 Mireille J
RESEARCH ARTICLE Neuropsychiatric Genetics Genetics of Aggressive Behavior: An Overview Kim Veroude,1 Yanli Zhang-James,2,3 Noelia Fernandez-Castillo,4,5,6 Mireille J. Bakker,1 Bru Cormand,4,5,6 and Stephen V. Faraone2,3,7* 1Department of Cognitive Neuroscience, Donders Institute for Brain, Cognition and Behaviour, Radboudumc, Nijmegen, The Netherlands 2Departments of Psychiatry and of Neuroscience and Physiology, SUNY Upstate Medical University, Syracuse, New York 3Departments of Neuroscience and Physiology, SUNY Upstate Medical University, Syracuse, New York 4Departament de Genetica, Facultat de Biologia, Universitat de Barcelona, Catalonia, Spain 5Institut de Biomedicina de la Universitat de Barcelona (IBUB), Catalonia, Spain 6Centro de Investigacion Biomedica en Red de Enfermedades Raras (CIBERER), Spain 7K.G. Jebsen Centre for Research on Neuropsychiatric Disorders, University of Bergen, Bergen, Norway Manuscript Received: 22 April 2015; Manuscript Accepted: 5 August 2015 The Research Domain Criteria (RDoC) address three types of aggression: frustrative non-reward, defensive aggression and How to Cite this Article: offensive/proactive aggression. This review sought to present Veroude K, Zhang-James Y, Fernandez- the evidence for genetic underpinnings of aggression and to Castillo N, Bakker MJ, Cormand B, determine to what degree prior studies have examined pheno- Faraone SV. 2016. Genetics of Aggressive types that fit into the RDoC framework. Although the constructs Behavior: An Overview. of defensive and offensive aggression have been widely used in the animal genetics literature, the human literature is mostly Am J Med Genet Part B 171B:3–43. agnostic with regard to all the RDoC constructs. We know from twin studies that about half the variance in behavior may be explained by genetic risk factors. -

A Web-Platform for Analysis of Host Factors Involved in Viral Infections Discovered by Genome Wide Rnai Screen
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is © The Royal Society of Chemistry 2017 vhfRNAi: A web-platform for analysis of host factors involved in viral infections discovered by genome wide RNAi screen Anamika Thakur#, Abid Qureshi# and Manoj Kumar* Bioinformatics Centre, Institute of Microbial Technology, Council of Scientific and Industrial Research, Sector 39A, Chandigarh-160036, India #Equal contribution * To whom correspondence should be addressed. Tel, 91-172-6665453; Fax, 91-172- 2690585; 91-172-2690632; Email, [email protected] Supplementary Tables Table S1: Statistics of unique and duplicate host factors in each virus Table S2: Table denoting genes common among different viruses Table S3: Statistics of GWAS analysis Table S1. Statistics of unique and duplicate host factors in each virus S. No. Virus Unique-Entries Duplicate-Entries 1. Adeno-associated virus (AAV) 926 533 2. Avian influenza virus (AIV) 0 11 3. Borna disease virus (BDV) 14 20 4. Dengue virus 2 (DEN-2) 27 13 5. Hepatitis C virus (HCV) 236 213 6. Human immunodeficiency virus 1 (HIV) 1388 857 7. Human parainfluenza virus 3 (HPIV-3) 0 27 8. Human herpesvirus 1 (HSV-1) 34 38 9. Influenza A virus (IAV) 700 513 10. Lymphocytic choriomeningitis virus (LCMV) 0 54 11. Marburgvirus (MARV) 0 11 12. Poliovirus (PV) 3340 1035 13. Rotavirus (RV) 347 175 14. Sendai virus (SeV) 32 27 15. Sindbis virus (SIV) 70 41 16. Vaccinia virus (VACV) 482 296 17. Vesicular stomatitis virus (VSV) 9 78 18. West Nile virus (WNV) 313 137 Table S1. Statistics of unique host factors for each virus having overlapping and unique factors in different viruses Non-overlap – Overall- S. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Genetics of Aggressive Behavior: an Overview
The influence of genes on “positive valence systems” constructs: A systematic review Item Type Article Authors Hess, Jonathan L.; Kawaguchi, Daniel M.; Wagner, Kayla E.; Faraone, Stephen V.; Glatt, Stephen J. Citation Hess, JL, Kawaguchi, DM, Wagner, KE, Faraone, SV, Glatt, SJ. 2015. The Influence of Genes on “Positive Valence Systems” Constructs: A Systematic Review. Am J Med Genet Part B 171B: 92– 110. DOI 10.1002/ajmg.b.32382 Publisher Wiley Rights Attribution-NonCommercial-NoDerivatives 4.0 International Download date 26/09/2021 01:12:21 Item License http://doi.wiley.com/10.1002/tdm_license_1.1 Link to Item http://hdl.handle.net/20.500.12648/1795 See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/281608624 Genetics of Aggressive Behavior: An Overview Article in American Journal of Medical Genetics Part B Neuropsychiatric Genetics · September 2015 DOI: 10.1002/ajmg.b.32364 CITATIONS READS 81 1,596 6 authors, including: Yanli Zhang-James Noèlia Fernàndez-Castillo State University of New York Upstate Medical University University of Barcelona 65 PUBLICATIONS 1,992 CITATIONS 102 PUBLICATIONS 848 CITATIONS SEE PROFILE SEE PROFILE Mireille J. Bakker Bru Cormand Radboud University Medical Centre (Radboudumc) University of Barcelona 8 PUBLICATIONS 272 CITATIONS 311 PUBLICATIONS 10,197 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Aggression Genetics View project Trastorno por deficit de atencion con hiperactividad en la clinica pediatrica View project All content following this page was uploaded by Noèlia Fernàndez-Castillo on 25 November 2017. The user has requested enhancement of the downloaded file. -

Characterization of Visceral and Subcutaneous Adipose Tissue
J. Perinat. Med. 2016; 44(7): 813–835 Shali Mazaki-Tovi*, Adi L. Tarca, Edi Vaisbuch, Juan Pedro Kusanovic, Nandor Gabor Than, Tinnakorn Chaiworapongsa, Zhong Dong, Sonia S. Hassan and Roberto Romero* Characterization of visceral and subcutaneous adipose tissue transcriptome in pregnant women with and without spontaneous labor at term: implication of alternative splicing in the metabolic adaptations of adipose tissue to parturition DOI 10.1515/jpm-2015-0259 groups (unpaired analyses) and adipose tissue regions Received July 27, 2015. Accepted October 26, 2015. Previously (paired analyses). Selected genes were tested by quantita- published online March 19, 2016. tive reverse transcription-polymerase chain reaction. Abstract Results: Four hundred and eighty-two genes were differ- entially expressed between visceral and subcutaneous Objective: The aim of this study was to determine gene fat of pregnant women with spontaneous labor at term expression and splicing changes associated with par- (q-value < 0.1; fold change > 1.5). Biological processes turition and regions (visceral vs. subcutaneous) of the enriched in this comparison included tissue and vascu- adipose tissue of pregnant women. lature development as well as inflammatory and meta- Study design: The transcriptome of visceral and abdomi- bolic pathways. Differential splicing was found for 42 nal subcutaneous adipose tissue from pregnant women at genes [q-value < 0.1; differences in Finding Isoforms using term with (n = 15) and without (n = 25) spontaneous labor Robust Multichip Analysis scores > 2] between adipose was profiled with the Affymetrix GeneChip Human Exon tissue regions of women not in labor. Differential exon 1.0 ST array. Overall gene expression changes and the dif- usage associated with parturition was found for three ferential exon usage rate were compared between patient genes (LIMS1, HSPA5, and GSTK1) in subcutaneous tissues. -
Figure S1. Reverse Transcription‑Quantitative PCR Analysis of ETV5 Mrna Expression Levels in Parental and ETV5 Stable Transfectants
Figure S1. Reverse transcription‑quantitative PCR analysis of ETV5 mRNA expression levels in parental and ETV5 stable transfectants. (A) Hec1a and Hec1a‑ETV5 EC cell lines; (B) Ishikawa and Ishikawa‑ETV5 EC cell lines. **P<0.005, unpaired Student's t‑test. EC, endometrial cancer; ETV5, ETS variant transcription factor 5. Figure S2. Survival analysis of sample clusters 1‑4. Kaplan Meier graphs for (A) recurrence‑free and (B) overall survival. Survival curves were constructed using the Kaplan‑Meier method, and differences between sample cluster curves were analyzed by log‑rank test. Figure S3. ROC analysis of hub genes. For each gene, ROC curve (left) and mRNA expression levels (right) in control (n=35) and tumor (n=545) samples from The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer cohort are shown. mRNA levels are expressed as Log2(x+1), where ‘x’ is the RSEM normalized expression value. ROC, receiver operating characteristic. Table SI. Clinicopathological characteristics of the GSE17025 dataset. Characteristic n % Atrophic endometrium 12 (postmenopausal) (Control group) Tumor stage I 91 100 Histology Endometrioid adenocarcinoma 79 86.81 Papillary serous 12 13.19 Histological grade Grade 1 30 32.97 Grade 2 36 39.56 Grade 3 25 27.47 Myometrial invasiona Superficial (<50%) 67 74.44 Deep (>50%) 23 25.56 aMyometrial invasion information was available for 90 of 91 tumor samples. Table SII. Clinicopathological characteristics of The Cancer Genome Atlas Uterine Corpus Endometrioid Cancer dataset. Characteristic n % Solid tissue normal 16 Tumor samples Stagea I 226 68.278 II 19 5.740 III 70 21.148 IV 16 4.834 Histology Endometrioid 271 81.381 Mixed 10 3.003 Serous 52 15.616 Histological grade Grade 1 78 23.423 Grade 2 91 27.327 Grade 3 164 49.249 Molecular subtypeb POLE 17 7.328 MSI 65 28.017 CN Low 90 38.793 CN High 60 25.862 CN, copy number; MSI, microsatellite instability; POLE, DNA polymerase ε. -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Resveratrol Inhibits Cell Cycle Progression by Targeting Aurora Kinase a and Polo-Like Kinase 1 in Breast Cancer Cells
3696 ONCOLOGY REPORTS 35: 3696-3704, 2016 Resveratrol inhibits cell cycle progression by targeting Aurora kinase A and Polo-like kinase 1 in breast cancer cells RUBICELI MEDINA-AGUILAR1, LAURENCE A. Marchat2, ELENA ARECHAGA OCAMPO3, Patricio GARIGLIO1, JAIME GARCÍA MENA1, NICOLÁS VILLEGAS SEPÚlveda4, MACARIO MartÍNEZ CASTILLO4 and CÉSAR LÓPEZ-CAMARILLO5 1Department of Genetics and Molecular Biology, CINVESTAV-IPN, Mexico D.F.; 2Molecular Biomedicine Program and Biotechnology Network, National School of Medicine and Homeopathy, National Polytechnic Institute, Mexico D.F.; 3Natural Sciences Department, Metropolitan Autonomous University, Mexico D.F.; 4Department of Molecular Biomedicine, CINVESTAV-IPN, Mexico D.F.; 5Oncogenomics and Cancer Proteomics Laboratory, Universidad Autónoma de la Ciudad de México, Mexico D.F., Mexico Received December 4, 2015; Accepted January 8, 2016 DOI: 10.3892/or.2016.4728 Abstract. The Aurora protein kinase (AURKA) and the MDA-MB-231 and MCF-7 cells. By western blot assays, we Polo-like kinase-1 (PLK1) activate the cell cycle, and they confirmed that resveratrol suppressed AURKA, CCND1 and are considered promising druggable targets in cancer CCNB1 at 24 and 48 h. In summary, we showed for the first time therapy. However, resistance to chemotherapy and to specific that resveratrol regulates cell cycle progression by targeting small-molecule inhibitors is common in cancer patients; thus AURKA and PLK1. Our findings highlight the potential use of alternative therapeutic approaches are needed to overcome resveratrol as an adjuvant therapy for breast cancer. clinical resistance. Here, we showed that the dietary compound resveratrol suppressed the cell cycle by targeting AURKA Introduction and PLK1 kinases. First, we identified genes modulated by resveratrol using a genome-wide analysis of gene expression Cancer development results from the interaction between in MDA-MB-231 breast cancer cells.