Transcriptomic Analysis of Early Stages of Intestinal Regeneration in Holothuria Glaberrima David J
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Critical Evaluation of Gene Expression Changes in Human Tissues In
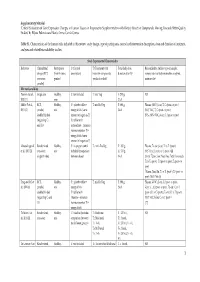
Supplementary Material ‘Critical Evaluation of Gene Expression Changes in Human Tissues in Response to Supplementation with Dietary Bioactive Compounds: Moving Towards Better-Quality Studies’ by Biljana Pokimica and María-Teresa García-Conesa Table S1. Characteristics of the human trials included in this review: study design, type of participants, control and intervention description, dose and duration of treatment, analyses and related bioavailability studies. Study Experimental Characteristics Reference Clinical trial Participants C (Control T (Treatment with Total daily dose, Bioavailability studies: type of sample, design (RCT, (health status, description) bioactive compounds, duration (d or h)1 compounds and (or) metabolites analysed, crossover, gender) products or diet) main results2 parallel) Mix meals and diets Persson I et al., Single arm Healthy, C: not included T: mix Veg T: 250 g, NR 2000 [1] men 21 d Møller P et al., RCT, Healthy, C1: placebo tablet + T: mix FruVeg T: 600 g, Plasma: (NS↑) β-car, T, C2 (post- vs pre-) 2003 [2] parallel, mix energy drink (same 24 d (NC) VitC, T, C2 (post- vs pre-) double blinded amount of sugars as T) (NS↓, 69%) VitC, β-car, C1 (post- vs pre-) (regarding C1 C2: tablet with and C2) antioxidants + minerals (same amount as T) + energy drink (same amount of sugars as T) Almendingen K Randomized, Healthy, C: no proper control T1,2: mix FruVeg T1: 300 g, Plasma: ↑α-car, β-car, T2 vs T1 (post-) et al., 2005 [3] crossover, mix included (comparison T2: 750 g, (NS↑) Lyc, Lut, T2 vs T1 (post-) [4] single -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

Smith Bacterial SBP56 Identified As a Cu-Dependent Methanethiol
Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere EYICE, Özge, MYRONOVA, Nataliia, POL, Arjan, CARRIÓN, Ornella, TODD, Jonathan D, SMITH, Thomas <http://orcid.org/0000-0002-4246-5020>, GURMAN, Stephen J, CUTHBERTSON, Adam, MAZARD, Sophie, MENNINK-KERSTEN, Monique Ash, BUGG, Timothy Dh, ANDERSSON, Karl Kristoffer, JOHNSTON, Andrew Wb, OP DEN CAMP, Huub Jm and SCHÄFER, Hendrik Available from Sheffield Hallam University Research Archive (SHURA) at: http://shura.shu.ac.uk/17252/ This document is the author deposited version. You are advised to consult the publisher's version if you wish to cite from it. Published version EYICE, Özge, MYRONOVA, Nataliia, POL, Arjan, CARRIÓN, Ornella, TODD, Jonathan D, SMITH, Thomas, GURMAN, Stephen J, CUTHBERTSON, Adam, MAZARD, Sophie, MENNINK-KERSTEN, Monique Ash, BUGG, Timothy Dh, ANDERSSON, Karl Kristoffer, JOHNSTON, Andrew Wb, OP DEN CAMP, Huub Jm and SCHÄFER, Hendrik (2018). Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. The ISME journal, 1 (12), 145-160. Copyright and re-use policy See http://shura.shu.ac.uk/information.html Sheffield Hallam University Research Archive http://shura.shu.ac.uk OPEN The ISME Journal (2017), 1–16 www.nature.com/ismej ORIGINAL ARTICLE Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere Özge Eyice1,2,9, Nataliia Myronova1,9, Arjan Pol3, Ornella Carrión4, Jonathan D Todd4, Tom J Smith5, Stephen J Gurman6, Adam Cuthbertson1, -

The Function of NM23-H1/NME1 and Its Homologs in Major Processes Linked to Metastasis
University of Dundee The Function of NM23-H1/NME1 and Its Homologs in Major Processes Linked to Metastasis Mátyási, Barbara; Farkas, Zsolt; Kopper, László; Sebestyén, Anna; Boissan, Mathieu; Mehta, Anil Published in: Pathology and Oncology Research DOI: 10.1007/s12253-020-00797-0 Publication date: 2020 Licence: CC BY Document Version Publisher's PDF, also known as Version of record Link to publication in Discovery Research Portal Citation for published version (APA): Mátyási, B., Farkas, Z., Kopper, L., Sebestyén, A., Boissan, M., Mehta, A., & Takács-Vellai, K. (2020). The Function of NM23-H1/NME1 and Its Homologs in Major Processes Linked to Metastasis. Pathology and Oncology Research, 26(1), 49-61. https://doi.org/10.1007/s12253-020-00797-0 General rights Copyright and moral rights for the publications made accessible in Discovery Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from Discovery Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain. • You may freely distribute the URL identifying the publication in the public portal. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Protein Targets of Acetaminophen Covalent Binding in Rat and Mouse
ORIGINAL RESEARCH published: XX XX 2021 doi: 10.3389/fchem.2021.736788 1 58 2 59 3 60 4 61 5 62 6 63 7 64 8 65 9 66 10 Protein Targets of Acetaminophen 67 11 68 12 Covalent Binding in Rat and Mouse 69 13 70 14 Q2 Liver Studied by LC-MS/MS 71 15 Q3 72 Q1 16 Timon Geib, Ghazaleh Moghaddam, Aimee Supinski, Makan Golizeh† and Lekha Sleno* Q4 73 17 Q5 74 18 Chemistry Department, Université du Québec à Montréal, Montréal, QC, Canada Q6 75 19 76 20 Acetaminophen (APAP) is a mild analgesic and antipyretic used commonly worldwide. 77 21 78 Although considered a safe and effective over-the-counter medication, it is also the leading 22 79 23 cause of drug-induced acute liver failure. Its hepatotoxicity has been linked to the covalent 80 24 binding of its reactive metabolite, N-acetyl p-benzoquinone imine (NAPQI), to proteins. The 81 Edited by: 25 aim of this study was to identify APAP-protein targets in both rat and mouse liver, and to 82 26 Marcus S Cooke, 83 University of South Florida, compare the results from both species, using bottom-up proteomics with data-dependent 27 United States 84 28 high resolution mass spectrometry and targeted multiple reaction monitoring (MRM) 85 Reviewed by: 29 experiments. Livers from rats and mice, treated with APAP, were homogenized and 86 Hartmut Jaeschke, 30 University of Kansas Medical Center digested by trypsin. Digests were then fractionated by mixed-mode solid-phase extraction 87 31 Research Institute, United States prior to liquid chromatography-tandem mass spectrometry (LC-MS/MS). -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota. -

Characterization of Visceral and Subcutaneous Adipose Tissue
J. Perinat. Med. 2016; 44(7): 813–835 Shali Mazaki-Tovi*, Adi L. Tarca, Edi Vaisbuch, Juan Pedro Kusanovic, Nandor Gabor Than, Tinnakorn Chaiworapongsa, Zhong Dong, Sonia S. Hassan and Roberto Romero* Characterization of visceral and subcutaneous adipose tissue transcriptome in pregnant women with and without spontaneous labor at term: implication of alternative splicing in the metabolic adaptations of adipose tissue to parturition DOI 10.1515/jpm-2015-0259 groups (unpaired analyses) and adipose tissue regions Received July 27, 2015. Accepted October 26, 2015. Previously (paired analyses). Selected genes were tested by quantita- published online March 19, 2016. tive reverse transcription-polymerase chain reaction. Abstract Results: Four hundred and eighty-two genes were differ- entially expressed between visceral and subcutaneous Objective: The aim of this study was to determine gene fat of pregnant women with spontaneous labor at term expression and splicing changes associated with par- (q-value < 0.1; fold change > 1.5). Biological processes turition and regions (visceral vs. subcutaneous) of the enriched in this comparison included tissue and vascu- adipose tissue of pregnant women. lature development as well as inflammatory and meta- Study design: The transcriptome of visceral and abdomi- bolic pathways. Differential splicing was found for 42 nal subcutaneous adipose tissue from pregnant women at genes [q-value < 0.1; differences in Finding Isoforms using term with (n = 15) and without (n = 25) spontaneous labor Robust Multichip Analysis scores > 2] between adipose was profiled with the Affymetrix GeneChip Human Exon tissue regions of women not in labor. Differential exon 1.0 ST array. Overall gene expression changes and the dif- usage associated with parturition was found for three ferential exon usage rate were compared between patient genes (LIMS1, HSPA5, and GSTK1) in subcutaneous tissues. -

Volatile Sulfur Compounds in Coastal Acid Sulfate Soils, Northern N.S.W
VOLATILE SULFUR COMPOUNDS IN COASTAL ACID SULFATE SOILS, NORTHERN N.S.W Andrew Stephen Kinsela A thesis submitted in fulfilment of the requirements for the degree of Doctor of Philosophy School of Biological, Earth & Environmental Sciences THE UNIVERSITY OF NEW SOUTH WALES, AUSTRALIA 2007 DECLARATION ORIGINALITY STATEMENT ‘I hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom I have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.’ Signed ………………………………………………… Date …………………………………………………… iii ACKNOWLEDGEMENTS There are numerous people who have assisted me throughout the course of my thesis. I therefore want to take this opportunity to thank a few of those who contributed appreciably, both directly and indirectly. First of all, I would like to express my heartfelt gratitude to my supervisor, Associate Professor Mike Melville. Mike’s initial teachings as part of my undergraduate studies first sparked my interest in soils. Since then his continued enthusiasm on the subject has helped shape the way I approach my own work. -

Development and Validation of a Protein-Based Risk Score for Cardiovascular Outcomes Among Patients with Stable Coronary Heart Disease
Supplementary Online Content Ganz P, Heidecker B, Hveem K, et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA. doi: 10.1001/jama.2016.5951 eTable 1. List of 1130 Proteins Measured by Somalogic’s Modified Aptamer-Based Proteomic Assay eTable 2. Coefficients for Weibull Recalibration Model Applied to 9-Protein Model eFigure 1. Median Protein Levels in Derivation and Validation Cohort eTable 3. Coefficients for the Recalibration Model Applied to Refit Framingham eFigure 2. Calibration Plots for the Refit Framingham Model eTable 4. List of 200 Proteins Associated With the Risk of MI, Stroke, Heart Failure, and Death eFigure 3. Hazard Ratios of Lasso Selected Proteins for Primary End Point of MI, Stroke, Heart Failure, and Death eFigure 4. 9-Protein Prognostic Model Hazard Ratios Adjusted for Framingham Variables eFigure 5. 9-Protein Risk Scores by Event Type This supplementary material has been provided by the authors to give readers additional information about their work. Downloaded From: https://jamanetwork.com/ on 10/02/2021 Supplemental Material Table of Contents 1 Study Design and Data Processing ......................................................................................................... 3 2 Table of 1130 Proteins Measured .......................................................................................................... 4 3 Variable Selection and Statistical Modeling ........................................................................................ -

Assessing the Human Canonical Protein Count[Version 1; Peer Review
F1000Research 2017, 6:448 Last updated: 15 JUL 2020 REVIEW Last rolls of the yoyo: Assessing the human canonical protein count [version 1; peer review: 1 approved, 2 approved with reservations] Christopher Southan IUPHAR/BPS Guide to Pharmacology, Centre for Integrative Physiology, University of Edinburgh, Edinburgh, EH8 9XD, UK First published: 07 Apr 2017, 6:448 Open Peer Review v1 https://doi.org/10.12688/f1000research.11119.1 Latest published: 07 Apr 2017, 6:448 https://doi.org/10.12688/f1000research.11119.1 Reviewer Status Abstract Invited Reviewers In 2004, when the protein estimate from the finished human genome was 1 2 3 only 24,000, the surprise was compounded as reviewed estimates fell to 19,000 by 2014. However, variability in the total canonical protein counts version 1 (i.e. excluding alternative splice forms) of open reading frames (ORFs) in 07 Apr 2017 report report report different annotation portals persists. This work assesses these differences and possible causes. A 16-year analysis of Ensembl and UniProtKB/Swiss-Prot shows convergence to a protein number of ~20,000. The former had shown some yo-yoing, but both have now plateaued. Nine 1 Michael Tress, Spanish National Cancer major annotation portals, reviewed at the beginning of 2017, gave a spread Research Centre (CNIO), Madrid, Spain of counts from 21,819 down to 18,891. The 4-way cross-reference concordance (within UniProt) between Ensembl, Swiss-Prot, Entrez Gene 2 Elspeth A. Bruford , European Molecular and the Human Gene Nomenclature Committee (HGNC) drops to 18,690, Biology Laboratory, Hinxton, UK indicating methodological differences in protein definitions and experimental existence support between sources.