Tumor SHB Gene Expression Affects Disease Characteristics in Human
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Supporting Information
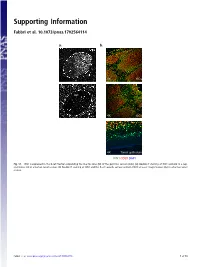
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

IBRAHIM ALDEAILEJ Phd.2013.Pdf
Bangor University DOCTOR OF PHILOSOPHY Identification and functional characterisation of germ line genes in human cancer cells Aldeailej, Ibrahim Award date: 2013 Awarding institution: Bangor University Link to publication General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 09. Oct. 2021 Bangor University School of Biological Sciences Identification and functional characterisation of germ line genes in human cancer cells Ph.D. Thesis 2013 IBRAHIM ALDEAILEJ Declaration and Consent Details of the Work I hereby agree to deposit the following item in the digital repository maintained by Bangor University and/or in any other repository authorized for use by Bangor University. Author Name: IBRAHIM MOHAMMED ALDEAILEJ Title: Identification and functional characterisation of germ line genes in human cancer cells Supervisor/Department: Dr. Ramsay James McFarlane/ School of Biological Sciences Funding body (if any): kingdom of Saudi Arabia Government Qualification/Degree obtained: Ph.D. -

Overexpression of the Proneural Transcription Factor ASCL1 In
Malli et al. Molecular Cytogenetics (2018) 11:3 DOI 10.1186/s13039-018-0355-7 CASE REPORT Open Access Overexpression of the proneural transcription factor ASCL1 in chronic lymphocytic leukemia with a t(12;14)(q23.2;q32.3) Theodora Malli1, Melanie Rammer1, Sabrina Haslinger1, Jonathan Burghofer1, Sonja Burgstaller2, Hans-Christian Boesmueller3,4, Renate Marschon1, Wolfgang Kranewitter1, Martin Erdel1, Sabine Deutschbauer1 and Gerald Webersinke1* Abstract Background: Translocations of the IGH locus on 14q32.3 are present in about 8% of patients with chronic lymphocytic leukemia (CLL) and contribute to leukemogenesis by deregulating the expression of the IGH-partner genes. Identification of these genes and investigation of the downstream effects of their deregulation can reveal disease-causing mechanisms. Case presentation: We report on the molecular characterization of a novel t(12;14)(q23.2;q32.3) in CLL. As a consequence of the rearrangement ASCL1 was brought into proximity of the IGHJ-Cμ enhancer and was highly overexpressed in the aberrant B-cells of the patient, as shown by qPCR and immunohistochemistry. ASCL1 encodes for a transcription factor acting as a master regulator of neurogenesis, is overexpressed in neuroendocrine tumors and a promising therapeutic target in small cell lung cancer (SCLC). Its overexpression has also been recently reported in acute adult T-cell leukemia/lymphoma. To examine possible downstream effects of the ASCL1 upregulation in CLL, we compared the gene expression of sorted CD5+ cells of the translocation patient to that of CD19+ B-cells from seven healthy donors and detected 176 significantly deregulated genes (Fold Change ≥2, FDR p ≤ 0.01). -

The Effect of Prpsc Accumulation on Inflammatory Gene
Edinburgh Research Explorer The effect of PrP(Sc) accumulation on inflammatory gene expression within sheep peripheral lymphoid tissue Citation for published version: Gossner, AG & Hopkins, J 2015, 'The effect of PrP(Sc) accumulation on inflammatory gene expression within sheep peripheral lymphoid tissue', Veterinary Microbiology, vol. 181, no. 3-4, pp. 204-211. https://doi.org/10.1016/j.vetmic.2015.10.013 Digital Object Identifier (DOI): 10.1016/j.vetmic.2015.10.013 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Veterinary Microbiology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 29. Sep. 2021 G Model VETMIC 7118 No. of Pages 8 Veterinary Microbiology xxx (2015) xxx–xxx Contents lists available at ScienceDirect Veterinary Microbiology journal homepage: www.elsevier.com/locate/vetmic Sc The effect of PrP accumulation on inflammatory gene expression within sheep peripheral lymphoid tissue Anton G. Gossner, John Hopkins* The Roslin Institute & R(D)SVS, University of Edinburgh, Easter Bush, Midlothian EH25 9RG, UK A R T I C L E I N F O A B S T R A C T Sc Article history: Accumulation of the misfolded prion protein, PrP in the central nervous system (CNS) is strongly linked Received 27 April 2015 to progressive neurodegenerative disease. -

Ep 2885427 B1
(19) TZZ _T (11) EP 2 885 427 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12Q 1/68 (2018.01) 10.01.2018 Bulletin 2018/02 (86) International application number: (21) Application number: 13759978.3 PCT/EP2013/002462 (22) Date of filing: 14.08.2013 (87) International publication number: WO 2014/026768 (20.02.2014 Gazette 2014/08) (54) COLORECTAL CANCER METHYLATION MARKER KOLOREKTALKREBSMETHYLIERUNGSMARKER MARQUEUR DE METHYLATION POUR LE CANCER COLORECTAL (84) Designated Contracting States: • RAFAEL A IRIZARRY ET AL: "The human colon AL AT BE BG CH CY CZ DE DK EE ES FI FR GB cancer methylome shows similar hypo- and GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO hypermethylation at conserved tissue-specific PL PT RO RS SE SI SK SM TR CpG island shores", NATURE GENETICS, vol. 41, no. 2, 1 February 2009 (2009-02-01), pages (30) Priority: 14.08.2012 EP 12180459 178-186, XP055029485, ISSN: 1061-4036, DOI: 10.1038/ng.298 (43) Date of publication of application: • M. D. ROBINSON ET AL: "Evaluation of 24.06.2015 Bulletin 2015/26 affinity-based genome-wide DNA methylation data: Effects of CpG density, amplification bias, (73) Proprietor: Max-Planck-Gesellschaft zur and copy number variation", GENOME Förderung RESEARCH, vol. 20, no. 12, 1 December 2010 der Wissenschaften (2010-12-01), pages 1719-1729, XP055041625, 80539 Munich (DE) ISSN: 1088-9051, DOI: 10.1101/gr.110601.110 • E. A. HOUSEMAN ET AL: "Copy number variation (72) Inventors: has little impact on bead-array-based measures • SCHWEIGER, Michal-Ruth of DNA methylation", BIOINFORMATICS, vol. -

The Convergent Roles of the Nuclear Factor I Transcription Factors in Development and Cancer
The convergent roles of the nuclear factor I transcription factors in development and cancer Kok-Siong Chen1, Jonathan W.C. Lim1, Linda J. Richards1,2*, Jens Bunt1* 1 Queensland Brain Institute, The University of Queensland, Brisbane, Australia 4072 2 The School of Biomedical Sciences, The University of Queensland, Brisbane, Australia 4072 * Corresponding authors: Jens Bunt: [email protected] and Linda J Richards: [email protected] 1 Highlights • NFI transcription factors play important roles in normal development and are associated with cancer in multiple organ systems. • NFI transcription factors are implicated in tumours that arise from cells that normally express the concordant transcription factor during development. • The role of NFI transcription factors in regulating the balance between cell proliferation and differentiation during development may be disrupted in cancer. • NFI transcription factors are context-dependent in various cancer types and can play either oncogenic or tumour-suppressive roles. 2 Abstract The nuclear factor I (NFI) transcription factors play important roles during normal development and have been associated with developmental abnormalities in humans. All four family members, NFIA, NFIB, NFIC and NFIX, have a homologous DNA binding domain and function by regulating cell proliferation and differentiation via the transcriptional control of their target genes. More recently, NFI genes have also been implicated in cancer based on genomic analyses and studies of animal models in a variety of tumours across multiple organ systems. However, the association between their functions in development and in cancer is not well described. In this review, we summarise the evidence suggesting a converging role for the NFI genes in development and cancer. -

Antibody Tools Immunohistochemistry
Antibody Tools Immunohistochemistry $$ 250 - 150 - 100 - 75 - 50 - 37 - Western Blot 25 - 20 - 15 - 10 - 1.4 1.2 1 0.8 0.6 OD 450 0.4 Sandwich ELISA 0.2 0 0.01 0.1 1 10 100 1000 Recombinant Protein Concentration(mg/ml) Immunohistochemistry Immunofluorescence 1 2 3 250 - 150 - 100 - 75 - 50 - Immunoprecipitation 37 - 25 - 20 - 15 - 100 80 60 % of Max 40 Flow Cytometry 20 0 3 4 5 0 102 10 10 10 www.abnova.com March 2013 (Sixth Edition) Abnova Corporation www.abnova.com Email: [email protected] Address: 9F, No. 108, Jhouzih St., Neihu, Taipei 114, Taiwan Tel: + 886 2 8751 1888 Fax: + 886 2 6602 1218 Antibodies tool for IHC, Class I IVD (In Vitro Diagnostics) Cat. Num. DH0003 DH0013 DH0020 DH0015 Product Name Anti-ACTN4 monoclonal antibody Anti-ANXA5 monoclonal antibody Anti-CDH17 monoclonal antibody Anti-CLDN1 monoclonal antibody Application Immunoperoxidase of monoclonal antibody Immunoperoxidase of monoclonal antibody to Immunoperoxidase of monoclonal antibody to Immunoperoxidase of monoclonal antibody to to ACTN4 on formalin-fixed paraffin- ANXA5 on formalin-fixed paraffin-embedded CDH17 on formalin-fixed paraffin-embedded CLDN1 on formalin-fixed paraffin-embedded embedded human pancreatic cancer. [antibody human colon cancer. [antibody concentration 3 human colon cancer. [antibody concentration 3 human colon cancer. [antibody concentration 3 concentration 1.5 ug/ml] ug/ml] ug/ml] ug/ml] Cat. Num. DH0002 DH0021 DH0010 DH0017 Product Name Anti-CTH monoclonal antibody Anti-EGR1 monoclonal antibody Anti-EIF2C2 monoclonal antibody Anti-ENO1 monoclonal antibody Application Immunoperoxidase of monoclonal antibody Immunoperoxidase of monoclonal antibody Immunoperoxidase of monoclonal antibody to Immunoperoxidase of monoclonal antibody to CTH on formalin-fixed paraffin-embedded to EGR1 on formalin-fixed paraffin-embedded EIF2C2 on formalin-fixed paraffin-embedded to ENO1 on formalin-fixed paraffin-embedded human liver. -

Whole Blood Gene Expression in Adolescent Chronic Fatigue Syndrome
Nguyen et al. J Transl Med (2017) 15:102 DOI 10.1186/s12967-017-1201-0 Journal of Translational Medicine RESEARCH Open Access Whole blood gene expression in adolescent chronic fatigue syndrome: an exploratory cross‑sectional study suggesting altered B cell diferentiation and survival Chinh Bkrong Nguyen1,2, Lene Alsøe3, Jessica M. Lindvall4, Dag Sulheim5, Even Fagermoen6, Anette Winger7, Mari Kaarbø8, Hilde Nilsen3 and Vegard Bruun Wyller1,2* Abstract Background: Chronic fatigue syndrome (CFS) is a prevalent and disabling condition afecting adolescents. The pathophysiology is poorly understood, but immune alterations might be an important component. This study compared whole blood gene expression in adolescent CFS patients and healthy controls, and explored associations between gene expression and neuroendocrine markers, immune markers and clinical markers within the CFS group. Methods: CFS patients (12–18 years old) were recruited nation-wide to a single referral center as part of the Nor- CAPITAL project. A broad case defnition of CFS was applied, requiring 3 months of unexplained, disabling chronic/ relapsing fatigue of new onset, whereas no accompanying symptoms were necessary. Healthy controls having comparable distribution of gender and age were recruited from local schools. Whole blood samples were subjected to RNA sequencing. Immune markers were blood leukocyte counts, plasma cytokines, serum C-reactive protein and immunoglobulins. Neuroendocrine markers encompassed plasma and urine levels of catecholamines and cortisol, as well as heart rate variability indices. Clinical markers consisted of questionnaire scores for symptoms of post-exertional malaise, infammation, fatigue, depression and trait anxiety, as well as activity recordings. Results: A total of 29 CFS patients and 18 healthy controls were included. -

Discovery of Rare Ancestry-Specific Variants in the Fetal Genome That Confer Risk of Preterm Premature Rupture of Membranes (PPROM) and Preterm Birth Bhavi P
Modi et al. BMC Medical Genetics (2018) 19:181 https://doi.org/10.1186/s12881-018-0696-4 TECHNICAL ADVANCE Open Access Discovery of rare ancestry-specific variants in the fetal genome that confer risk of preterm premature rupture of membranes (PPROM) and preterm birth Bhavi P. Modi1, Hardik I. Parikh2, Maria E. Teves3, Rewa Kulkarni1, Jiang Liyu3, Roberto Romero4,5, Timothy P. York1,3 and Jerome F. Strauss III1,3* Abstract Background: Preterm premature rupture of membranes (PPROM) is the leading identifiable cause of preterm birth, a complication that is more common in African Americans. Attempts to identify genetic loci associated with preterm birth using genome-wide association studies (GWAS) have only been successful with large numbers of cases and controls, and there has yet to be a convincing genetic association to explain racial/ethnic disparities. Indeed, the search for ancestry-specific variants associated with preterm birth has led to the conclusion that spontaneous preterm birth could be the consequence of multiple rare variants. The hypothesis that preterm birth is due to rare genetic variants that would go undetected in standard GWAS has been explored in the present study. The detection and validation of these rare variants present challenges because of the low allele frequency. However, some success in the identification of fetal loci/genes associated with preterm birth using whole genome sequencing and whole exome sequencing (WES) has recently been reported. While encouraging, this is currently an expensive technology, and methods to leverage the sequencing data to quickly identify and cost-effectively validate variants are needed. Methods: We developed a WES data analysis strategy based on neonatal genomic DNA from PPROM cases and term controls that was unencumbered by preselection of candidate genes, and capable of identifying variants in African Americans worthy of focused evaluation to establish statistically significant associations. -

Transcriptional Co-Regulation of Micrornas and Protein-Coding Genes
Transcriptional co-regulation of microRNAs and protein-coding genes A thesis submitted to the University of Manchester for the degree of Doctor of Philosophy in the Faculty of Life Sciences 2013 By Aaron Webber Table of Contents Abstract ......................................................................................................................................... 8 Declaration .................................................................................................................................... 9 Copyright statement ................................................................................................................... 10 Acknowledgements ..................................................................................................................... 11 Preface ........................................................................................................................................ 12 Chapter 1: Introduction .............................................................................................................. 13 1.1 The protein-coding gene expression pathway ............................................................ 13 1.2 Regulation of transcription ......................................................................................... 17 1.2.1 Transcription factors ........................................................................................... 19 1.2.2 Transcriptional regulatory regions ...................................................................... 22 -

Short Course I
SHORT COURSE I Using iPS Cells and Reprogramming to Model Neural Development and Disease Organized by Kevin Eggan, PhD Short Course I Using iPS Cells and Reprogramming to Model Neural Development and Disease Organized by Kevin Eggan, PhD Please cite articles using the model: [AUTHOR’S LAST NAME, AUTHOR’S FIRST & MIDDLE INITIALS] (2015) [CHAPTER TITLE] In: Using iPS Cells and Reprogramming to Model Neural Development and Disease. (Eggan K, ed) pp. [xx-xx]. Chicago, IL: Society for Neuroscience. All articles and their graphics are under the copyright of their respective authors. Cover graphics and design © 2015 Society for Neuroscience. Table of Contents Probing Disorders of the Nervous System Using Reprogramming Approaches Evangelos Kiskinis, PhD . 7 Stem Cells As a Tool for Studying the Developmental Regulation of Gene Expression Hynek Wichterle, PhD, Michael Closser, BS, Esteban O . Mazzoni, PhD, Shaun Mahony, PhD, Yuchun Guo, PhD, Rachel Kopunova, and David K . Gifford, PhD . 17 Generating a Functional Human Cortex In Vitro From Induced Pluripotent Stem Cells Sergiu P . Pas¸ca, MD . 27 Generating 3D Cerebral Organoids From Human Pluripotent Stem Cells to Model Cortical Development and Disease Paola Arlotta, PhD . 41 Drug-Based Modulation of Endogenous Stem Cells Promotes Functional Remyelination In Vivo Fadi J . Najm, MBA, Mayur Madhavan, PhD, Anita Zaremba, BA, Elizabeth Shick, BS, Robert T . Karl, BS, Daniel C . Factor, BA, Tyler E . Miller, BS, Zachary S . Nevin, BS, Christopher Kantor, Alex Sargent, Kevin L . Quick, Daniela M . Schlatzer, Hong Tang, Ruben Papoian, PhD, Kyle R . Brimacombe, MS, Min Shen, Matthew B . Boxer, Ajit Jadhav, Andrew P . Robinson, Joseph R . -

Identification and Mechanistic Investigation of Recurrent Functional Genomic and Transcriptional Alterations in Advanced Prostat
Identification and Mechanistic Investigation of Recurrent Functional Genomic and Transcriptional Alterations in Advanced Prostate Cancer Thomas A. White A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Washington 2013 Reading Committee: Peter S. Nelson, Chair Janet L. Stanford Raymond J. Monnat Jay A. Shendure Program Authorized to Offer Degree: Molecular and Cellular Biology ©Copyright 2013 Thomas A. White University of Washington Abstract Identification and Mechanistic Investigation of Recurrent Functional Genomic and Transcriptional Alterations in Advanced Prostate Cancer Thomas A. White Chair of the Supervisory Committee: Peter S. Nelson, MD Novel functionally altered transcripts may be recurrent in prostate cancer (PCa) and may underlie lethal and advanced disease and the neuroendocrine small cell carcinoma (SCC) phenotype. We conducted an RNASeq survey of the LuCaP series of 24 PCa xenograft tumors from 19 men, and validated observations on metastatic tumors and PCa cell lines. Key findings include discovery and validation of 40 novel fusion transcripts including one recurrent chimera, many SCC-specific and castration resistance (CR) -specific novel splice isoforms, new observations on SCC-specific and TMPRSS2-ERG specific differential expression, the allele-specific expression of certain recurrent non- synonymous somatic single nucleotide variants (nsSNVs) previously discovered via exome sequencing of the same tumors, rgw ubiquitous A-to-I RNA editing of base excision repair (BER) gene NEIL1 as well as CDK13, a kinase involved in RNA splicing, and the SCC expression of a previously unannotated long noncoding RNA (lncRNA) at Chr6p22.2. Mechanistic investigation of the novel lncRNA indicates expression is regulated by derepression by master neuroendocrine regulator RE1-silencing transcription factor (REST), and may regulate some genes of axonogenesis and angiogenesis.