The Effect of Prpsc Accumulation on Inflammatory Gene
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of Novel Pathways That Promote Anoikis Through Genome-Wide Screens
University of Massachusetts Medical School eScholarship@UMMS GSBS Dissertations and Theses Graduate School of Biomedical Sciences 2016-10-14 Identification of Novel Pathways that Promote Anoikis through Genome-wide Screens Victoria E. Pedanou University of Massachusetts Medical School Let us know how access to this document benefits ou.y Follow this and additional works at: https://escholarship.umassmed.edu/gsbs_diss Part of the Biology Commons, and the Cancer Biology Commons Repository Citation Pedanou VE. (2016). Identification of Novel Pathways that Promote Anoikis through Genome-wide Screens. GSBS Dissertations and Theses. https://doi.org/10.13028/M27G6D. Retrieved from https://escholarship.umassmed.edu/gsbs_diss/889 This material is brought to you by eScholarship@UMMS. It has been accepted for inclusion in GSBS Dissertations and Theses by an authorized administrator of eScholarship@UMMS. For more information, please contact [email protected]. i TITLE PAGE IDENTIFICATION OF NOVEL PATHWAYS THAT PROMOTE ANOIKIS THROUGH GENOME-WIDE SCREENS A Dissertation Presented By VICTORIA ELIZABETH PEDANOU Submitted to the Faculty of the University of Massachusetts Graduate School of Biomedical Sciences, Worcester in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY OCTOBER 14TH, 2016 CANCER BIOLOGY ii SIGNATURE PAGE IDENTIFICATION OF NOVEL PATHWAYS THAT PROMOTE ANOIKIS THROUGH GENOME-WIDE SCREENS A Dissertation Presented By VICTORIA ELIZABETH PEDANOU This work was undertaken in the Graduate School of Biomedical Sciences Cancer Biology The signature of the Thesis Advisor signifies validation of Dissertation content ___________________________ Michael R. Green, Thesis Advisor The signatures of the Dissertation Defense Committee signify completion and approval as to style and content of the Dissertation __________________________________ Eric H. -
Supporting Information
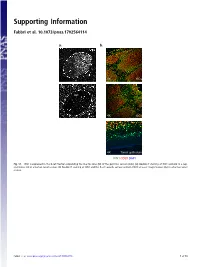
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

The Correlation of Keratin Expression with In-Vitro Epithelial Cell Line Differentiation
The correlation of keratin expression with in-vitro epithelial cell line differentiation Deeqo Aden Thesis submitted to the University of London for Degree of Master of Philosophy (MPhil) Supervisors: Professor Ian. C. Mackenzie Professor Farida Fortune Centre for Clinical and Diagnostic Oral Science Barts and The London School of Medicine and Dentistry Queen Mary, University of London 2009 Contents Content pages ……………………………………………………………………......2 Abstract………………………………………………………………………….........6 Acknowledgements and Declaration……………………………………………...…7 List of Figures…………………………………………………………………………8 List of Tables………………………………………………………………………...12 Abbreviations….………………………………………………………………..…...14 Chapter 1: Literature review 16 1.1 Structure and function of the Oral Mucosa……………..…………….…..............17 1.2 Maintenance of the oral cavity...……………………………………….................20 1.2.1 Environmental Factors which damage the Oral Mucosa………. ….…………..21 1.3 Structure and function of the Oral Mucosa ………………...….……….………...21 1.3.1 Skin Barrier Formation………………………………………………….……...22 1.4 Comparison of Oral Mucosa and Skin…………………………………….……...24 1.5 Developmental and Experimental Models used in Oral mucosa and Skin...……..28 1.6 Keratinocytes…………………………………………………….….....................29 1.6.1 Desmosomes…………………………………………….…...............................29 1.6.2 Hemidesmosomes……………………………………….…...............................30 1.6.3 Tight Junctions………………………….……………….…...............................32 1.6.4 Gap Junctions………………………….……………….….................................32 -

IBRAHIM ALDEAILEJ Phd.2013.Pdf
Bangor University DOCTOR OF PHILOSOPHY Identification and functional characterisation of germ line genes in human cancer cells Aldeailej, Ibrahim Award date: 2013 Awarding institution: Bangor University Link to publication General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 09. Oct. 2021 Bangor University School of Biological Sciences Identification and functional characterisation of germ line genes in human cancer cells Ph.D. Thesis 2013 IBRAHIM ALDEAILEJ Declaration and Consent Details of the Work I hereby agree to deposit the following item in the digital repository maintained by Bangor University and/or in any other repository authorized for use by Bangor University. Author Name: IBRAHIM MOHAMMED ALDEAILEJ Title: Identification and functional characterisation of germ line genes in human cancer cells Supervisor/Department: Dr. Ramsay James McFarlane/ School of Biological Sciences Funding body (if any): kingdom of Saudi Arabia Government Qualification/Degree obtained: Ph.D. -

MALE Protein Name Accession Number Molecular Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean H Mean PDAC Mean T-Test PDAC Vs. H T-Test
MALE t-test t-test Accession Molecular H PDAC PDAC vs. PDAC vs. Protein Name Number Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean Mean Mean H CP PDAC/H PDAC/CP - 22 kDa protein IPI00219910 22 kDa 7 5 4 8 1 0 6 6 1 0.1126 0.0456 0.1 0.1 - Cold agglutinin FS-1 L-chain (Fragment) IPI00827773 12 kDa 32 39 34 26 53 57 36 30 55 0.0309 0.0388 1.8 1.5 - HRV Fab 027-VL (Fragment) IPI00827643 12 kDa 4 6 0 0 0 0 5 0 0 - 0.0574 - 0.0 - REV25-2 (Fragment) IPI00816794 15 kDa 8 12 5 7 8 9 10 6 8 0.2225 0.3844 1.3 0.8 A1BG Alpha-1B-glycoprotein precursor IPI00022895 54 kDa 115 109 106 112 111 100 112 109 105 0.6497 0.4138 1.0 0.9 A2M Alpha-2-macroglobulin precursor IPI00478003 163 kDa 62 63 86 72 14 18 63 79 16 0.0120 0.0019 0.2 0.3 ABCB1 Multidrug resistance protein 1 IPI00027481 141 kDa 41 46 23 26 52 64 43 25 58 0.0355 0.1660 2.4 1.3 ABHD14B Isoform 1 of Abhydrolase domain-containing proteinIPI00063827 14B 22 kDa 19 15 19 17 15 9 17 18 12 0.2502 0.3306 0.7 0.7 ABP1 Isoform 1 of Amiloride-sensitive amine oxidase [copper-containing]IPI00020982 precursor85 kDa 1 5 8 8 0 0 3 8 0 0.0001 0.2445 0.0 0.0 ACAN aggrecan isoform 2 precursor IPI00027377 250 kDa 38 30 17 28 34 24 34 22 29 0.4877 0.5109 1.3 0.8 ACE Isoform Somatic-1 of Angiotensin-converting enzyme, somaticIPI00437751 isoform precursor150 kDa 48 34 67 56 28 38 41 61 33 0.0600 0.4301 0.5 0.8 ACE2 Isoform 1 of Angiotensin-converting enzyme 2 precursorIPI00465187 92 kDa 11 16 20 30 4 5 13 25 5 0.0557 0.0847 0.2 0.4 ACO1 Cytoplasmic aconitate hydratase IPI00008485 98 kDa 2 2 0 0 0 0 2 0 0 - 0.0081 - 0.0 -

Transcriptome Profiling and Differential Gene Expression In
G C A T T A C G G C A T genes Article Transcriptome Profiling and Differential Gene Expression in Canine Microdissected Anagen and Telogen Hair Follicles and Interfollicular Epidermis Dominique J. Wiener 1,* ,Kátia R. Groch 1 , Magdalena A.T. Brunner 2,3, Tosso Leeb 2,3 , Vidhya Jagannathan 2 and Monika M. Welle 3,4 1 Department of Veterinary Pathobiology, College of Veterinary Medicine & Biomedical Science, Texas A&M University, College Station, TX 77843, USA; [email protected] 2 Institute of Genetics, Vetsuisse Faculty, University of Bern, 3012 Bern, Switzerland; [email protected] (M.A.T.B.); [email protected] (T.L.); [email protected] (V.J.) 3 Dermfocus, Vetsuisse Faculty, University Hospital of Bern, 3010 Bern, Switzerland; [email protected] 4 Institute of Animal Pathology, Vetsuisse Faculty, University of Bern, 3012 Bern, Switzerland * Correspondence: [email protected]; Tel.: +1-979-862-1568 Received: 30 June 2020; Accepted: 3 August 2020; Published: 4 August 2020 Abstract: The transcriptome profile and differential gene expression in telogen and late anagen microdissected hair follicles and the interfollicular epidermis of healthy dogs was investigated by using RNAseq. The genes with the highest expression levels in each group were identified and genes known from studies in other species to be associated with structure and function of hair follicles and epidermis were evaluated. Transcriptome profiling revealed that late anagen follicles expressed mainly keratins and telogen follicles expressed GSN and KRT15. The interfollicular epidermis expressed predominately genes encoding for proteins associated with differentiation. All sample groups express genes encoding for proteins involved in cellular growth and signal transduction. -

Damage of Hair Follicle Stem Cells and Alteration of Keratin Expression in External Radiation-Induced Acute Alopecia
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 30: 579-584, 2012 Damage of hair follicle stem cells and alteration of keratin expression in external radiation-induced acute alopecia NAOKI NANASHIMA, KOICHI ITO, TAKASHI ISHIKAWA, MANABU NAKANO and TOSHIYA NAKAMURA Department of Biomedical Sciences, Division of Medical Life Sciences, Hirosaki University Graduate School of Health Sciences, Hirosaki, Japan Received April 4, 2012; Accepted May 28, 2012 DOI: 10.3892/ijmm.2012.1018 Abstract. Alopecia is known as a symptom of acute radia- disturbances and blood and bone marrow disorders are known tion, yet little is known concerning the mechanism of this to occur within several hours to several weeks after 1-6 Gy of phenomenon and the alteration of hair protein profiles. To radiation exposure (4,6). examine this, 6-week-old male C57/BL6 mice were exposed Hair loss is also an effect of ARS, but little is known to 6 Gy of X-ray irradiation, which caused acute alopecia. about the mechanism underlying radiation-induced hair loss. Their hair and skin were collected, and hair proteins were In humans, hair loss is caused by radiation of more than analyzed with liquid chromatography/electrospray-ionization 3 Gy, and almost complete hair loss occurs within weeks of mass spectrometry and immunohistochemistry. No change exposure to 6 Gy (4,6). Since blood stem cells are sensitive to was observed in the composition of major hair keratins, such radiation (7), hair loss is thought to be caused by irradiation- as Krt81, Krt83 and Krt86. However, cytokeratin Krt15 and induced stem cell damage, yet no studies have investigated CD34, which are known as hair follicle stem cell markers, this hypothesis. -

Overexpression of the Proneural Transcription Factor ASCL1 In
Malli et al. Molecular Cytogenetics (2018) 11:3 DOI 10.1186/s13039-018-0355-7 CASE REPORT Open Access Overexpression of the proneural transcription factor ASCL1 in chronic lymphocytic leukemia with a t(12;14)(q23.2;q32.3) Theodora Malli1, Melanie Rammer1, Sabrina Haslinger1, Jonathan Burghofer1, Sonja Burgstaller2, Hans-Christian Boesmueller3,4, Renate Marschon1, Wolfgang Kranewitter1, Martin Erdel1, Sabine Deutschbauer1 and Gerald Webersinke1* Abstract Background: Translocations of the IGH locus on 14q32.3 are present in about 8% of patients with chronic lymphocytic leukemia (CLL) and contribute to leukemogenesis by deregulating the expression of the IGH-partner genes. Identification of these genes and investigation of the downstream effects of their deregulation can reveal disease-causing mechanisms. Case presentation: We report on the molecular characterization of a novel t(12;14)(q23.2;q32.3) in CLL. As a consequence of the rearrangement ASCL1 was brought into proximity of the IGHJ-Cμ enhancer and was highly overexpressed in the aberrant B-cells of the patient, as shown by qPCR and immunohistochemistry. ASCL1 encodes for a transcription factor acting as a master regulator of neurogenesis, is overexpressed in neuroendocrine tumors and a promising therapeutic target in small cell lung cancer (SCLC). Its overexpression has also been recently reported in acute adult T-cell leukemia/lymphoma. To examine possible downstream effects of the ASCL1 upregulation in CLL, we compared the gene expression of sorted CD5+ cells of the translocation patient to that of CD19+ B-cells from seven healthy donors and detected 176 significantly deregulated genes (Fold Change ≥2, FDR p ≤ 0.01). -

WO 2013/184908 A2 12 December 2013 (12.12.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2013/184908 A2 12 December 2013 (12.12.2013) P O P C T (51) International Patent Classification: Jr.; One Procter & Gamble Plaza, Cincinnati, Ohio 45202 G06F 19/00 (201 1.01) (US). HOWARD, Brian, Wilson; One Procter & Gamble Plaza, Cincinnati, Ohio 45202 (US). (21) International Application Number: PCT/US20 13/044497 (74) Agents: GUFFEY, Timothy, B. et al; c/o The Procter & Gamble Company, Global Patent Services, 299 East 6th (22) Date: International Filing Street, Sycamore Building, 4th Floor, Cincinnati, Ohio 6 June 2013 (06.06.2013) 45202 (US). (25) Filing Language: English (81) Designated States (unless otherwise indicated, for every (26) Publication Language: English kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, (30) Priority Data: BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, 61/656,218 6 June 2012 (06.06.2012) US DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (71) Applicant: THE PROCTER & GAMBLE COMPANY HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KN, KP, KR, [US/US]; One Procter & Gamble Plaza, Cincinnati, Ohio KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, ME, 45202 (US). MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SC, (72) Inventors: XU, Jun; One Procter & Gamble Plaza, Cincin SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, nati, Ohio 45202 (US). -

Types I and II Keratin Intermediate Filaments
Downloaded from http://cshperspectives.cshlp.org/ on October 10, 2021 - Published by Cold Spring Harbor Laboratory Press Types I and II Keratin Intermediate Filaments Justin T. Jacob,1 Pierre A. Coulombe,1,2 Raymond Kwan,3 and M. Bishr Omary3,4 1Department of Biochemistry and Molecular Biology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, Maryland 21205 2Departments of Biological Chemistry, Dermatology, and Oncology, School of Medicine, and Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University, Baltimore, Maryland 21205 3Departments of Molecular & Integrative Physiologyand Medicine, Universityof Michigan, Ann Arbor, Michigan 48109 4VA Ann Arbor Health Care System, Ann Arbor, Michigan 48105 Correspondence: [email protected] SUMMARY Keratins—types I and II—are the intermediate-filament-forming proteins expressed in epithe- lial cells. They are encoded by 54 evolutionarily conserved genes (28 type I, 26 type II) and regulated in a pairwise and tissue type–, differentiation-, and context-dependent manner. Here, we review how keratins serve multiple homeostatic and stress-triggered mechanical and nonmechanical functions, including maintenance of cellular integrity, regulation of cell growth and migration, and protection from apoptosis. These functions are tightly regulated by posttranslational modifications and keratin-associated proteins. Genetically determined alterations in keratin-coding sequences underlie highly penetrant and rare disorders whose pathophysiology reflects cell fragility or altered -

Ep 2885427 B1
(19) TZZ _T (11) EP 2 885 427 B1 (12) EUROPEAN PATENT SPECIFICATION (45) Date of publication and mention (51) Int Cl.: of the grant of the patent: C12Q 1/68 (2018.01) 10.01.2018 Bulletin 2018/02 (86) International application number: (21) Application number: 13759978.3 PCT/EP2013/002462 (22) Date of filing: 14.08.2013 (87) International publication number: WO 2014/026768 (20.02.2014 Gazette 2014/08) (54) COLORECTAL CANCER METHYLATION MARKER KOLOREKTALKREBSMETHYLIERUNGSMARKER MARQUEUR DE METHYLATION POUR LE CANCER COLORECTAL (84) Designated Contracting States: • RAFAEL A IRIZARRY ET AL: "The human colon AL AT BE BG CH CY CZ DE DK EE ES FI FR GB cancer methylome shows similar hypo- and GR HR HU IE IS IT LI LT LU LV MC MK MT NL NO hypermethylation at conserved tissue-specific PL PT RO RS SE SI SK SM TR CpG island shores", NATURE GENETICS, vol. 41, no. 2, 1 February 2009 (2009-02-01), pages (30) Priority: 14.08.2012 EP 12180459 178-186, XP055029485, ISSN: 1061-4036, DOI: 10.1038/ng.298 (43) Date of publication of application: • M. D. ROBINSON ET AL: "Evaluation of 24.06.2015 Bulletin 2015/26 affinity-based genome-wide DNA methylation data: Effects of CpG density, amplification bias, (73) Proprietor: Max-Planck-Gesellschaft zur and copy number variation", GENOME Förderung RESEARCH, vol. 20, no. 12, 1 December 2010 der Wissenschaften (2010-12-01), pages 1719-1729, XP055041625, 80539 Munich (DE) ISSN: 1088-9051, DOI: 10.1101/gr.110601.110 • E. A. HOUSEMAN ET AL: "Copy number variation (72) Inventors: has little impact on bead-array-based measures • SCHWEIGER, Michal-Ruth of DNA methylation", BIOINFORMATICS, vol. -

Identification of Novel Wool Keratin Intermediate Filament Genes in Sheep Skin Z-D
222 Yu et al. – Wool keratin genes Identification of novel wool keratin intermediate filament genes in sheep skin Z-D. YU1*, S.W. GORDON1, 2, J.E. WILDERMOTH1, O.A.M. WALLACE1, A.J. NIXON1 and A.J. PEARSON1 1AgResearch Ruakura, Private Bag 3123, Hamilton 3240, New Zealand 2Excerpta Medica, Sing Pao Building New Wing, 101 King's Road, North Point, Hong Kong *Corresponding author: [email protected] ABSTRACT Keratin intermediate filament (KIF) genes encode key proteins for hair and wool formation. A total of 17 expressed human KIF genes involved in hair formation are annotated. However, to date, only eight wool KIF genes have been reported. In this study, six new cDNA sequences (KRT32, KRT33B, KRT34, KRT39, KRT40 and KRT82) representing previously unreported wool KIFs were identified using a contiguous ovine sequence library constructed primarily from ESTs. The expression of three other KIF genes (KRT36, KRT84 and KRT87) was confirmed by PCR using sheep skin total RNA. The analogue of human KRT37 (type I) was unable to be identified, while KRT87 (type II) was present in sheep but not in humans. Therefore 10 type I and seven type II KIF family members have been identified in sheep in comparison to 11 and six KIFs in the human. These 17 KIF genes are likely to represent the complete or near-complete set involved in wool formation. The annotation of these genes will facilitate investigation into their patterns of expression in wool follicles and their roles in the determination of fibre attributes. Key words: keratin intermediate filament; wool; gene expression.