Association of KRT35, KRT2.10 and BFMS Polymorphisms with Wool Quality Traits in Egyptian Sheep Breeds
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of Novel Pathways That Promote Anoikis Through Genome-Wide Screens
University of Massachusetts Medical School eScholarship@UMMS GSBS Dissertations and Theses Graduate School of Biomedical Sciences 2016-10-14 Identification of Novel Pathways that Promote Anoikis through Genome-wide Screens Victoria E. Pedanou University of Massachusetts Medical School Let us know how access to this document benefits ou.y Follow this and additional works at: https://escholarship.umassmed.edu/gsbs_diss Part of the Biology Commons, and the Cancer Biology Commons Repository Citation Pedanou VE. (2016). Identification of Novel Pathways that Promote Anoikis through Genome-wide Screens. GSBS Dissertations and Theses. https://doi.org/10.13028/M27G6D. Retrieved from https://escholarship.umassmed.edu/gsbs_diss/889 This material is brought to you by eScholarship@UMMS. It has been accepted for inclusion in GSBS Dissertations and Theses by an authorized administrator of eScholarship@UMMS. For more information, please contact [email protected]. i TITLE PAGE IDENTIFICATION OF NOVEL PATHWAYS THAT PROMOTE ANOIKIS THROUGH GENOME-WIDE SCREENS A Dissertation Presented By VICTORIA ELIZABETH PEDANOU Submitted to the Faculty of the University of Massachusetts Graduate School of Biomedical Sciences, Worcester in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY OCTOBER 14TH, 2016 CANCER BIOLOGY ii SIGNATURE PAGE IDENTIFICATION OF NOVEL PATHWAYS THAT PROMOTE ANOIKIS THROUGH GENOME-WIDE SCREENS A Dissertation Presented By VICTORIA ELIZABETH PEDANOU This work was undertaken in the Graduate School of Biomedical Sciences Cancer Biology The signature of the Thesis Advisor signifies validation of Dissertation content ___________________________ Michael R. Green, Thesis Advisor The signatures of the Dissertation Defense Committee signify completion and approval as to style and content of the Dissertation __________________________________ Eric H. -

75 2. INTRODUCTION Triple-Negative Breast Cancer (TNBC)
[Frontiers in Bioscience, Scholar, 11, 75-88, March 1, 2019] The persisting puzzle of racial disparity in triple negative breast cancer: looking through a new lens Chakravarthy Garlapati1, Shriya Joshi1, Bikram Sahoo1, Shobhna Kapoor2, Ritu Aneja1 1Department of Biology, Georgia State University, Atlanta, GA, USA, 2Department of Chemistry, Indian Institute of Technology Bombay, Powai, India TABLE OF CONTENTS 1. Abstract 2. Introduction 3. Dissecting the TNBC racially disparate burden 3.1. Does race influence TNBC onset and progression? 3.2. Tumor microenvironment in TNBC and racial disparity 3.3. Differential gene signatures and pathways in racially distinct TNBC 3.4. Our Perspective: Looking racial disparity through a new lens 4. Conclusion 5. Acknowledgement 6. References 1. ABSTRACT 2. INTRODUCTION Triple-negative breast cancer (TNBC) Triple-negative breast cancer (TNBC), is characterized by the absence of estrogen a subtype of breast cancer (BC), accounts for and progesterone receptors and absence 15-20% of all BC diagnoses in the US. It has of amplification of human epidermal growth been recognized that women of African descent factor receptor (HER2). This disease has no are twice as likely to develop TNBC than approved treatment with a poor prognosis women of European descent (1). As the name particularly in African-American (AA) as foretells, TNBCs lack estrogen, progesterone, compared to European-American (EA) and human epidermal growth factor receptors. patients. Gene ontology analysis showed Unfortunately, TNBCs are defined by what they specific gene pathways that are differentially “lack” rather than what they “have” and thus this regulated and gene signatures that are negative nomenclature provides no actionable differentially expressed in AA as compared to information on “druggable” targets. -

The Correlation of Keratin Expression with In-Vitro Epithelial Cell Line Differentiation
The correlation of keratin expression with in-vitro epithelial cell line differentiation Deeqo Aden Thesis submitted to the University of London for Degree of Master of Philosophy (MPhil) Supervisors: Professor Ian. C. Mackenzie Professor Farida Fortune Centre for Clinical and Diagnostic Oral Science Barts and The London School of Medicine and Dentistry Queen Mary, University of London 2009 Contents Content pages ……………………………………………………………………......2 Abstract………………………………………………………………………….........6 Acknowledgements and Declaration……………………………………………...…7 List of Figures…………………………………………………………………………8 List of Tables………………………………………………………………………...12 Abbreviations….………………………………………………………………..…...14 Chapter 1: Literature review 16 1.1 Structure and function of the Oral Mucosa……………..…………….…..............17 1.2 Maintenance of the oral cavity...……………………………………….................20 1.2.1 Environmental Factors which damage the Oral Mucosa………. ….…………..21 1.3 Structure and function of the Oral Mucosa ………………...….……….………...21 1.3.1 Skin Barrier Formation………………………………………………….……...22 1.4 Comparison of Oral Mucosa and Skin…………………………………….……...24 1.5 Developmental and Experimental Models used in Oral mucosa and Skin...……..28 1.6 Keratinocytes…………………………………………………….….....................29 1.6.1 Desmosomes…………………………………………….…...............................29 1.6.2 Hemidesmosomes……………………………………….…...............................30 1.6.3 Tight Junctions………………………….……………….…...............................32 1.6.4 Gap Junctions………………………….……………….….................................32 -

Appendix 2. Significantly Differentially Regulated Genes in Term Compared with Second Trimester Amniotic Fluid Supernatant
Appendix 2. Significantly Differentially Regulated Genes in Term Compared With Second Trimester Amniotic Fluid Supernatant Fold Change in term vs second trimester Amniotic Affymetrix Duplicate Fluid Probe ID probes Symbol Entrez Gene Name 1019.9 217059_at D MUC7 mucin 7, secreted 424.5 211735_x_at D SFTPC surfactant protein C 416.2 206835_at STATH statherin 363.4 214387_x_at D SFTPC surfactant protein C 295.5 205982_x_at D SFTPC surfactant protein C 288.7 1553454_at RPTN repetin solute carrier family 34 (sodium 251.3 204124_at SLC34A2 phosphate), member 2 238.9 206786_at HTN3 histatin 3 161.5 220191_at GKN1 gastrokine 1 152.7 223678_s_at D SFTPA2 surfactant protein A2 130.9 207430_s_at D MSMB microseminoprotein, beta- 99.0 214199_at SFTPD surfactant protein D major histocompatibility complex, class II, 96.5 210982_s_at D HLA-DRA DR alpha 96.5 221133_s_at D CLDN18 claudin 18 94.4 238222_at GKN2 gastrokine 2 93.7 1557961_s_at D LOC100127983 uncharacterized LOC100127983 93.1 229584_at LRRK2 leucine-rich repeat kinase 2 HOXD cluster antisense RNA 1 (non- 88.6 242042_s_at D HOXD-AS1 protein coding) 86.0 205569_at LAMP3 lysosomal-associated membrane protein 3 85.4 232698_at BPIFB2 BPI fold containing family B, member 2 84.4 205979_at SCGB2A1 secretoglobin, family 2A, member 1 84.3 230469_at RTKN2 rhotekin 2 82.2 204130_at HSD11B2 hydroxysteroid (11-beta) dehydrogenase 2 81.9 222242_s_at KLK5 kallikrein-related peptidase 5 77.0 237281_at AKAP14 A kinase (PRKA) anchor protein 14 76.7 1553602_at MUCL1 mucin-like 1 76.3 216359_at D MUC7 mucin 7, -

MALE Protein Name Accession Number Molecular Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean H Mean PDAC Mean T-Test PDAC Vs. H T-Test
MALE t-test t-test Accession Molecular H PDAC PDAC vs. PDAC vs. Protein Name Number Weight CP1 CP2 H1 H2 PDAC1 PDAC2 CP Mean Mean Mean H CP PDAC/H PDAC/CP - 22 kDa protein IPI00219910 22 kDa 7 5 4 8 1 0 6 6 1 0.1126 0.0456 0.1 0.1 - Cold agglutinin FS-1 L-chain (Fragment) IPI00827773 12 kDa 32 39 34 26 53 57 36 30 55 0.0309 0.0388 1.8 1.5 - HRV Fab 027-VL (Fragment) IPI00827643 12 kDa 4 6 0 0 0 0 5 0 0 - 0.0574 - 0.0 - REV25-2 (Fragment) IPI00816794 15 kDa 8 12 5 7 8 9 10 6 8 0.2225 0.3844 1.3 0.8 A1BG Alpha-1B-glycoprotein precursor IPI00022895 54 kDa 115 109 106 112 111 100 112 109 105 0.6497 0.4138 1.0 0.9 A2M Alpha-2-macroglobulin precursor IPI00478003 163 kDa 62 63 86 72 14 18 63 79 16 0.0120 0.0019 0.2 0.3 ABCB1 Multidrug resistance protein 1 IPI00027481 141 kDa 41 46 23 26 52 64 43 25 58 0.0355 0.1660 2.4 1.3 ABHD14B Isoform 1 of Abhydrolase domain-containing proteinIPI00063827 14B 22 kDa 19 15 19 17 15 9 17 18 12 0.2502 0.3306 0.7 0.7 ABP1 Isoform 1 of Amiloride-sensitive amine oxidase [copper-containing]IPI00020982 precursor85 kDa 1 5 8 8 0 0 3 8 0 0.0001 0.2445 0.0 0.0 ACAN aggrecan isoform 2 precursor IPI00027377 250 kDa 38 30 17 28 34 24 34 22 29 0.4877 0.5109 1.3 0.8 ACE Isoform Somatic-1 of Angiotensin-converting enzyme, somaticIPI00437751 isoform precursor150 kDa 48 34 67 56 28 38 41 61 33 0.0600 0.4301 0.5 0.8 ACE2 Isoform 1 of Angiotensin-converting enzyme 2 precursorIPI00465187 92 kDa 11 16 20 30 4 5 13 25 5 0.0557 0.0847 0.2 0.4 ACO1 Cytoplasmic aconitate hydratase IPI00008485 98 kDa 2 2 0 0 0 0 2 0 0 - 0.0081 - 0.0 -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Transcriptome Profiling and Differential Gene Expression In
G C A T T A C G G C A T genes Article Transcriptome Profiling and Differential Gene Expression in Canine Microdissected Anagen and Telogen Hair Follicles and Interfollicular Epidermis Dominique J. Wiener 1,* ,Kátia R. Groch 1 , Magdalena A.T. Brunner 2,3, Tosso Leeb 2,3 , Vidhya Jagannathan 2 and Monika M. Welle 3,4 1 Department of Veterinary Pathobiology, College of Veterinary Medicine & Biomedical Science, Texas A&M University, College Station, TX 77843, USA; [email protected] 2 Institute of Genetics, Vetsuisse Faculty, University of Bern, 3012 Bern, Switzerland; [email protected] (M.A.T.B.); [email protected] (T.L.); [email protected] (V.J.) 3 Dermfocus, Vetsuisse Faculty, University Hospital of Bern, 3010 Bern, Switzerland; [email protected] 4 Institute of Animal Pathology, Vetsuisse Faculty, University of Bern, 3012 Bern, Switzerland * Correspondence: [email protected]; Tel.: +1-979-862-1568 Received: 30 June 2020; Accepted: 3 August 2020; Published: 4 August 2020 Abstract: The transcriptome profile and differential gene expression in telogen and late anagen microdissected hair follicles and the interfollicular epidermis of healthy dogs was investigated by using RNAseq. The genes with the highest expression levels in each group were identified and genes known from studies in other species to be associated with structure and function of hair follicles and epidermis were evaluated. Transcriptome profiling revealed that late anagen follicles expressed mainly keratins and telogen follicles expressed GSN and KRT15. The interfollicular epidermis expressed predominately genes encoding for proteins associated with differentiation. All sample groups express genes encoding for proteins involved in cellular growth and signal transduction. -

Damage of Hair Follicle Stem Cells and Alteration of Keratin Expression in External Radiation-Induced Acute Alopecia
INTERNATIONAL JOURNAL OF MOLECULAR MEDICINE 30: 579-584, 2012 Damage of hair follicle stem cells and alteration of keratin expression in external radiation-induced acute alopecia NAOKI NANASHIMA, KOICHI ITO, TAKASHI ISHIKAWA, MANABU NAKANO and TOSHIYA NAKAMURA Department of Biomedical Sciences, Division of Medical Life Sciences, Hirosaki University Graduate School of Health Sciences, Hirosaki, Japan Received April 4, 2012; Accepted May 28, 2012 DOI: 10.3892/ijmm.2012.1018 Abstract. Alopecia is known as a symptom of acute radia- disturbances and blood and bone marrow disorders are known tion, yet little is known concerning the mechanism of this to occur within several hours to several weeks after 1-6 Gy of phenomenon and the alteration of hair protein profiles. To radiation exposure (4,6). examine this, 6-week-old male C57/BL6 mice were exposed Hair loss is also an effect of ARS, but little is known to 6 Gy of X-ray irradiation, which caused acute alopecia. about the mechanism underlying radiation-induced hair loss. Their hair and skin were collected, and hair proteins were In humans, hair loss is caused by radiation of more than analyzed with liquid chromatography/electrospray-ionization 3 Gy, and almost complete hair loss occurs within weeks of mass spectrometry and immunohistochemistry. No change exposure to 6 Gy (4,6). Since blood stem cells are sensitive to was observed in the composition of major hair keratins, such radiation (7), hair loss is thought to be caused by irradiation- as Krt81, Krt83 and Krt86. However, cytokeratin Krt15 and induced stem cell damage, yet no studies have investigated CD34, which are known as hair follicle stem cell markers, this hypothesis. -

The Effect of Prpsc Accumulation on Inflammatory Gene
Edinburgh Research Explorer The effect of PrP(Sc) accumulation on inflammatory gene expression within sheep peripheral lymphoid tissue Citation for published version: Gossner, AG & Hopkins, J 2015, 'The effect of PrP(Sc) accumulation on inflammatory gene expression within sheep peripheral lymphoid tissue', Veterinary Microbiology, vol. 181, no. 3-4, pp. 204-211. https://doi.org/10.1016/j.vetmic.2015.10.013 Digital Object Identifier (DOI): 10.1016/j.vetmic.2015.10.013 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: Veterinary Microbiology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 29. Sep. 2021 G Model VETMIC 7118 No. of Pages 8 Veterinary Microbiology xxx (2015) xxx–xxx Contents lists available at ScienceDirect Veterinary Microbiology journal homepage: www.elsevier.com/locate/vetmic Sc The effect of PrP accumulation on inflammatory gene expression within sheep peripheral lymphoid tissue Anton G. Gossner, John Hopkins* The Roslin Institute & R(D)SVS, University of Edinburgh, Easter Bush, Midlothian EH25 9RG, UK A R T I C L E I N F O A B S T R A C T Sc Article history: Accumulation of the misfolded prion protein, PrP in the central nervous system (CNS) is strongly linked Received 27 April 2015 to progressive neurodegenerative disease. -

A Mixture of Tocopherol Acetate and L-Menthol Synergistically Promotes Hair Growth in C57BL/6 Mice
pharmaceutics Article A Mixture of Tocopherol Acetate and L-Menthol Synergistically Promotes Hair Growth in C57BL/6 Mice Seunghyun Ahn , Jung Yeon Lee, Sang Mi Choi, Yujeong Shin and Seyeon Park * Department of Applied Chemistry, Dongduk Women’s University, Seoul 02748, Korea; [email protected] (S.A.); [email protected] (J.Y.L.); [email protected] (S.M.C.); [email protected] (Y.S.) * Correspondence: [email protected]; Tel.: +82-02-940-4514 Received: 4 November 2020; Accepted: 15 December 2020; Published: 18 December 2020 Abstract: Oral finasteride and topical minoxidil are single components approved by the US FDA for treating hair loss. Some other compounds originating from natural products are also traditionally used for promoting hair growth. In this study, observations of treated keratinocyte cells were used to demonstrate that tocopherol acetate, L-menthol, and stevioside exert an effect on cell regeneration. Furthermore, these were topically applied to the shaved skin of C57BL/6 mice to observe their effects on hair growth. A mixture of tocopherol acetate, L-menthol, and stevioside showed the highest potential for promoting hair growth in vivo. In in vivo experiments, the mixture of tocopherol acetate, L-menthol, and stevioside was more effective than tocopherol acetate or L-menthol alone in promoting hair growth. The transcriptome analysis of skin from the dorsal side of a mouse treated with tocopherol acetate or L-menthol versus vehicle revealed key changes in keratin, keratin-associated protein, forkhead box, sonic hedgehog, fibroblast growth factor 10, desmoglein 4, deoxyribonuclease 1-like 2, and cadherin 3, known to play roles in promoting hair growth. -
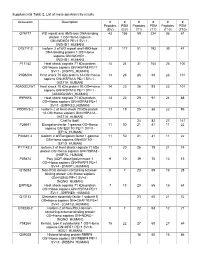
Supplemental Table 2: List of Mass-Spectrometry Results
Supplemental Table 2: List of mass-spectrometry results Accession Description # # # # # # Peptides PSM Peptides PSM Peptides PSM (EV) (EV) (T1) (T1) (T10) (T10) O75717 WD repeat and HMG-box DNA-binding 43 155 59 224 36 61 protein 1 OS=Homo sapiens GN=WDHD1 PE=1 SV=1 - [WDHD1_HUMAN] O75717-2 Isoform 2 of WD repeat and HMG-box 37 117 51 192 30 47 DNA-binding protein 1 OS=Homo sapiens GN=WDHD1 - [WDHD1_HUMAN] P11142 Heat shock cognate 71 kDa protein 14 24 31 104 26 100 OS=Homo sapiens GN=HSPA8 PE=1 SV=1 - [HSP7C_HUMAN] P0DMV8 Heat shock 70 kDa protein 1A OS=Homo 14 23 26 93 23 101 sapiens GN=HSPA1A PE=1 SV=1 - [HS71A_HUMAN] A0A0G2JIW1 Heat shock 70 kDa protein 1B OS=Homo 14 23 26 93 23 101 sapiens GN=HSPA1B PE=1 SV=1 - [A0A0G2JIW1_HUMAN] E9PKE3 Heat shock cognate 71 kDa protein 13 22 29 91 24 88 OS=Homo sapiens GN=HSPA8 PE=1 SV=1 - [E9PKE3_HUMAN] P0DMV8-2 Isoform 2 of Heat shock 70 kDa protein 13 19 25 86 22 95 1A OS=Homo sapiens GN=HSPA1A - [HS71A_HUMAN] Chaf1a (bait) 24 82 27 141 P26641 Elongation factor 1-gamma OS=Homo 11 50 21 81 11 22 sapiens GN=EEF1G PE=1 SV=3 - [EF1G_HUMAN] P26641-2 Isoform 2 of Elongation factor 1-gamma 11 50 21 81 11 22 OS=Homo sapiens GN=EEF1G - [EF1G_HUMAN] P11142-2 Isoform 2 of Heat shock cognate 71 kDa 11 20 26 79 21 74 protein OS=Homo sapiens GN=HSPA8 - [HSP7C_HUMAN] P09874 Poly [ADP-ribose] polymerase 1 9 10 39 70 13 15 OS=Homo sapiens GN=PARP1 PE=1 SV=4 - [PARP1_HUMAN] Q15233 Non-POU domain-containing octamer- 5 7 23 69 13 28 binding protein OS=Homo sapiens GN=NONO PE=1 SV=4 - [NONO_HUMAN] E9PNE6 Heat shock -

Analyses of Histological and Transcriptome Differences in the Skin
Ding et al. BMC Genomics (2019) 20:140 https://doi.org/10.1186/s12864-019-5503-x RESEARCH ARTICLE Open Access Analyses of histological and transcriptome differences in the skin of short-hair and long-hair rabbits Haisheng Ding, Huiling Zhao, Guanglong Cheng, Yongxin Yang, Xiaofei Wang, Xiaowei Zhao, Yunxia Qi and Dongwei Huang* Abstract Background: Hair fibre length is an important economic trait of rabbits in fur production. However, molecular mechanisms regulating rabbit hair growth have remained elusive. Results: Here we aimed to characterise the skin traits and gene expression profiles of short-hair and long-hair rabbits by histological and transcriptome analyses. Haematoxylin-eosin staining was performed to observe the histological structure of the skin of short-hair and long-hair rabbits. Compared to that in short-hair rabbits, a significantly longer anagen phase was observed in long-hair rabbits. In addition, by RNA sequencing, we identified 951 genes that were expressed at significantly different levels in the skin of short-hair and long-hair rabbits. Nine significantly differentially expressed genes were validated by quantitative real-time polymerase chain reaction. A gene ontology analysis revealed that epidermis development, hair follicle development, and lipid metabolic process were significantly enriched. Further, we identified potential functional genes regulating follicle development, lipid metabolic, and apoptosis as well as important pathways including extracellular matrix-receptor interaction and basal cell carcinoma pathway. Conclusions: The present study provides transcriptome evidence for the differences in hair growth between short- hair and long-hair rabbits and reveals that lipid metabolism and apoptosis might constitute major factors contributing to hair length.