The Correlation of Keratin Expression with In-Vitro Epithelial Cell Line Differentiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Development and Maintenance of Epidermal Stem Cells in Skin Adnexa
International Journal of Molecular Sciences Review Development and Maintenance of Epidermal Stem Cells in Skin Adnexa Jaroslav Mokry * and Rishikaysh Pisal Medical Faculty, Charles University, 500 03 Hradec Kralove, Czech Republic; [email protected] * Correspondence: [email protected] Received: 30 October 2020; Accepted: 18 December 2020; Published: 20 December 2020 Abstract: The skin surface is modified by numerous appendages. These structures arise from epithelial stem cells (SCs) through the induction of epidermal placodes as a result of local signalling interplay with mesenchymal cells based on the Wnt–(Dkk4)–Eda–Shh cascade. Slight modifications of the cascade, with the participation of antagonistic signalling, decide whether multipotent epidermal SCs develop in interfollicular epidermis, scales, hair/feather follicles, nails or skin glands. This review describes the roles of epidermal SCs in the development of skin adnexa and interfollicular epidermis, as well as their maintenance. Each skin structure arises from distinct pools of epidermal SCs that are harboured in specific but different niches that control SC behaviour. Such relationships explain differences in marker and gene expression patterns between particular SC subsets. The activity of well-compartmentalized epidermal SCs is orchestrated with that of other skin cells not only along the hair cycle but also in the course of skin regeneration following injury. This review highlights several membrane markers, cytoplasmic proteins and transcription factors associated with epidermal SCs. Keywords: stem cell; epidermal placode; skin adnexa; signalling; hair pigmentation; markers; keratins 1. Epidermal Stem Cells as Units of Development 1.1. Development of the Epidermis and Placode Formation The embryonic skin at very early stages of development is covered by a surface ectoderm that is a precursor to the epidermis and its multiple derivatives. -

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Inhibiting TNIK for Treating Colon Cancer
(19) & (11) EP 2 305 717 A1 (12) EUROPEAN PATENT APPLICATION (43) Date of publication: (51) Int Cl.: 06.04.2011 Bulletin 2011/14 C07K 16/40 (2006.01) C12N 15/11 (2006.01) C12Q 1/48 (2006.01) C12Q 1/68 (2006.01) (2006.01) (21) Application number: 09170853.7 G01N 33/50 (22) Date of filing: 21.09.2009 (84) Designated Contracting States: • Mahmoudi, Tokameh AT BE BG CH CY CZ DE DK EE ES FI FR GB GR 3515 XS, Utrecht (NL) HR HU IE IS IT LI LT LU LV MC MK MT NL NO PL • Clevers, Johannes Carolus PT RO SE SI SK SM TR 3712 AP, Huis ter Heide (NL) (71) Applicant: KoninklijkeNederlandse Akademie van (74) Representative: Swinkels, Bart Willem Wetenschappen Nederlandsch Octrooibureau 1011 JV Amsterdam (NL) J. W. Frisolaan 13 2517 JS Den Haag (NL) (72) Inventors: • Wing Li, Vivian Sze 3572 SH, Utrecht (NL) (54) Inhibiting TNIK for treating colon cancer (57) The invention relates to an inhibitor of TNIK and its use for treating cancer. EP 2 305 717 A1 Printed by Jouve, 75001 PARIS (FR) EP 2 305 717 A1 Description Field of the invention 5 [0001] The invention relates to an inhibitor of TNIK and its use as a medicament for treating cancer. Background of the invention [0002] The primary function of the intestinal tract involves the digestion and absorption of nutrients. The intestinal 10 lumen is lined with a specialized simple epithelium, which performs the primary functions of digestion, water and nutrient absorption and forms a barrier against luminal pathogens. -

Associated Palmoplantar Keratoderma
DR ABIGAIL ZIEMAN (Orcid ID : 0000-0001-8236-207X) Article type : Review Article Pathophysiology of pachyonychia congenita-associated palmoplantar keratoderma: New insight into skin epithelial homeostasis and avenues for treatment Authors: A. G. Zieman1 and P. A. Coulombe1,2 # Affiliations: 1Department of Cell and Developmental Biology, University of Michigan Medical School, Ann Arbor, MI 48109, USA; 2Department of Dermatology, University of Michigan Medical School, Ann Arbor, MI 48109, USA #Corresponding author: Pierre A. Coulombe, PhD, 3071 Biomedical Sciences Research Building, 109 Zina Pitcher Place, Ann Arbor, MI 48109, USA. Tel: 734-615-7509. Email: [email protected]. Funding Sources: These studies were supported by grant AR044232 issued to P.A.C. from the National Institute of Arthritis, Musculoskeletal and Skin Disease (NIAMS). A.G.Z. received support from grant T32 CA009110 from the National Cancer Institute. Author Manuscript This is the author manuscript accepted for publication and has undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/BJD.18033 This article is protected by copyright. All rights reserved Conflict of interest disclosures: None declared. Bulleted statements: What’s already known about this topic? Pachyonychia congenita is a rare genodermatosis caused by mutations in KRT6A, KRT6B, KRT6C, KRT16, KRT17, which are normally expressed in skin appendages and induced following injury. Individuals with PC present with multiple clinical symptoms that usually include thickened and dystrophic nails, palmoplantar keratoderma (PPK), glandular cysts, and oral leukokeratosis. -

Transgenic Cyclooxygenase-2 Overexpression Sensitizes Mouse Skin for Carcinogenesis
Transgenic cyclooxygenase-2 overexpression sensitizes mouse skin for carcinogenesis Karin Mu¨ ller-Decker*†, Gitta Neufang*, Irina Berger‡, Melanie Neumann*, Friedrich Marks*, and Gerhard Fu¨ rstenberger* *Research Program Tumor Cell Regulation, Deutsches Krebsforschungszentrum, and ‡Department of Pathology, Ruprecht-Karls-University, 69120 Heidelberg, Germany Edited by Philip Needleman, Pharmacia Corporation, St. Louis, MO, and approved July 29, 2002 (received for review May 30, 2002) Genetic and pharmacological evidence suggests that overexpres- there is a causal relationship between COX-2 overexpression sion of cyclooxygenase-2 (COX-2) is critical for epithelial carcino- and tumor development. Recently, we have shown that the genesis and provides a major target for cancer chemoprevention keratin 5 (K5) promoter-driven overexpression of COX-2 in by nonsteroidal antiinflammatory drugs. Transgenic mouse lines basal cells of interfollicular epidermis and the pilosebaceous unit with keratin 5 promoter-driven COX-2 overexpression in basal led to a preneoplastic skin phenotype in 4 of 4 high-expression epidermal cells exhibit a preneoplastic skin phenotype. As shown mouse lines (15). here, this phenotype depends on the level of COX-2 expression and To delineate COX-2 functions for carcinogenesis, we have COX-2-mediated prostaglandin accumulation. The transgenics did used the initiation–promotion model (2) for the induction of skin not develop skin tumors spontaneously but did so after a single tumors in wild-type (wt) NMRI mice and COX-2 transgenic application of an initiating dose of the carcinogen 7,12-dimethyl- mouse lines. This multistage model allows the analysis of the benz[a]anthracene (DMBA). Long-term treatment with the tumor carcinogenic process in terms of distinct stages, i.e., initiation by promoter phorbol 12-myristate 13-acetate, as required for tumor- application of a subcarcinogenic dose of a carcinogen such as igenesis in wild-type mice, was not necessary for transgenics. -

Molecular Pathways Associated with the Nutritional Programming of Plant-Based Diet Acceptance in Rainbow Trout Following an Early Feeding Exposure Mukundh N
Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, Aurélie Le Cam, Françoise Médale, Sadasivam J. Kaushik, Inge Geurden To cite this version: Mukundh N. Balasubramanian, Stéphane Panserat, Mathilde Dupont-Nivet, Edwige Quillet, Jérôme Montfort, et al.. Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure. BMC Genomics, BioMed Central, 2016, 17 (1), pp.1-20. 10.1186/s12864-016-2804-1. hal-01346912 HAL Id: hal-01346912 https://hal.archives-ouvertes.fr/hal-01346912 Submitted on 19 Jul 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Balasubramanian et al. BMC Genomics (2016) 17:449 DOI 10.1186/s12864-016-2804-1 RESEARCH ARTICLE Open Access Molecular pathways associated with the nutritional programming of plant-based diet acceptance in rainbow trout following an early feeding exposure Mukundh N. Balasubramanian1, Stephane Panserat1, Mathilde Dupont-Nivet2, Edwige Quillet2, Jerome Montfort3, Aurelie Le Cam3, Francoise Medale1, Sadasivam J. Kaushik1 and Inge Geurden1* Abstract Background: The achievement of sustainable feeding practices in aquaculture by reducing the reliance on wild-captured fish, via replacement of fish-based feed with plant-based feed, is impeded by the poor growth response seen in fish fed high levels of plant ingredients. -
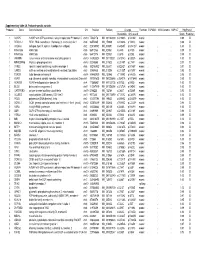
Supplementary Table 2A. Proband-Specific Variants Proband
Supplementary table 2A. Proband-specific variants Proband Gene Gene full name Chr Position RefSeqChange Function ESP6500 1000 Genome SNP137 PolyPhen2 Nucleotide Amino acid Score Prediction 1 AGAP5 ArfGAP with GTPase domain, ankyrin repeat and PH domain 5 chr10 75435178 NM_001144000 c.G1240A p.V414M exonic . 0.99 D 1 REXO1L1 REX1, RNA exonuclease 1 homolog (S. cerevisiae)-like 1 chr8 86574465 NM_172239 c.A1262G p.Y421C exonic . 0.98 D 1 COL4A3 collagen, type IV, alpha 3 (Goodpasture antigen) chr2 228169782 NM_000091 c.G4235T p.G1412V exonic . 1.00 D 1 KIAA1586 KIAA1586 chr6 56912133 NM_020931 c.C44G p.A15G exonic . 0.99 D 1 KIAA1586 KIAA1586 chr6 56912176 NM_020931 c.C87G p.D29E exonic . 0.99 D 1 JAKMIP3 Janus kinase and microtubule interacting protein 3 chr10 133955524 NM_001105521 c.G1574C p.G525A exonic . 1.00 D 1 MPHOSPH8 M-phase phosphoprotein 8 chr13 20240685 NM_017520 c.C2140T p.L714F exonic . 0.97 D 1 SYNE1 spectrin repeat containing, nuclear envelope 1 chr6 152783902 NM_033071 c.A2242T p.N748Y exonic . 0.97 D 1 CAND2 cullin-associated and neddylation-dissociated 2 (putative) chr3 12858835 NM_012298 c.C2125T p.H709Y exonic . 0.95 D 1 TDRD9 tudor domain containing 9 chr14 104460909 NM_153046 c.T1289G p.V430G exonic . 0.96 D 2 ASPM asp (abnormal spindle) homolog, microcephaly associated (Drosophila)chr1 197091625 NM_001206846 c.G3491A p.R1164H exonic . 0.99 D 2 ADAM29 ADAM metallopeptidase domain 29 chr4 175896951 NM_001130705 c.A275G p.D92G exonic . 1.00 D 2 BCO2 beta-carotene oxygenase 2 chr11 112087019 NM_001256398 c.G1373A p.G458E exonic . -

Hypomesus Transpacificus
Aquatic Toxicology 105 (2011) 369–377 Contents lists available at ScienceDirect Aquatic Toxicology jou rnal homepage: www.elsevier.com/locate/aquatox Sublethal responses to ammonia exposure in the endangered delta smelt; Hypomesus transpacificus (Fam. Osmeridae) ∗ 1 2 Richard E. Connon , Linda A. Deanovic, Erika B. Fritsch, Leandro S. D’Abronzo , Inge Werner Aquatic Toxicology Laboratory, Department of Anatomy, Physiology and Cell Biology, School of Veterinary Medicine, University of California, Davis, California 95616, United States a r t i c l e i n f o a b s t r a c t Article history: The delta smelt (Hypomesus transpacificus) is an endangered pelagic fish species endemic to the Received 9 May 2011 Sacramento-San Joaquin Estuary in Northern California, which acts as an indicator of ecosystem health Received in revised form 29 June 2011 in its habitat range. Interrogative tools are required to successfully monitor effects of contaminants upon Accepted 2 July 2011 the delta smelt, and to research potential causes of population decline in this species. We used microarray technology to investigate genome-wide effects in fish exposed to ammonia; one of multiple contami- Keywords: nants arising from wastewater treatment plants and agricultural runoff. A 4-day exposure of 57-day Hypomesus transpacificus + old juveniles resulted in a total ammonium (NH4 –N) median lethal concentration (LC50) of 13 mg/L, Delta smelt Microarray and a corresponding un-ionized ammonia (NH3) LC50 of 147 g/L. Using the previously designed delta + Biomarker smelt microarray we assessed altered gene transcription in juveniles exposed to 10 mg/L NH4 –N from Ammonia this 4-day exposure. -

Keratin 9 Point Mutation in the Pedigree of Epidermolytic Hereditary Palmoplantar Keratoderma Perturbs Keratin Intermediate Filament Network Formation
FEBS 17004 FEBS Letters 386 (1996) 149-155 Keratin 9 point mutation in the pedigree of epidermolytic hereditary palmoplantar keratoderma perturbs keratin intermediate filament network formation Setsu Kobayashi, Toshihiro Tanaka*, Norihisa Matsuyoshi, Sadao Imamura Department of Dermatology, Graduate School of Medicine, Kyoto University, Kyoto, 606 Japan Received 12 January 1996; revised version received 4 April 1996 Abstract Keratins form an intracellular keratin filament net- point mutations in the K9 gene in EHPPK [4-8] but none work in keratinocytes. Point mutations in the epidermal keratins showed a function assay with these mutations. Here, we pro- could lead to the disruption of keratin filament formation, vide the first demonstration that the point mutation found in developing skin diseases such as epidermolytic hereditary a pedigree of EHPPK has a dominant-negative effect on the palmoplantar keratoderma (EHPPK). We found a G to A assembly of keratin intermediate filaments in the cells. transition in keratin 9 (K9) cDNA, resulting in the substitution of glutamine for arginine at 162, in all patients of a pedigree of 2. Materials and methods EHPPK. Transfection into MDCK cells and DJM-1 cells revealed that the plasmid CMX vector containing normal keratin 2.1. PCR and DNA sequence 9 cDNA showed normal keratin network formation, whereas the Genomic DNA was extracted and purified from blood or biopsy vector with a G to A point mutated keratin 9 cDNA showed specimens from the patients. The primers were designed at nucleotide disrupted keratin filaments with droplet formation in the cells. 263 282 and 664-683 based on the K9 cDNA sequence [9]. -

Cellular and Molecular Signatures in the Disease Tissue of Early
Cellular and Molecular Signatures in the Disease Tissue of Early Rheumatoid Arthritis Stratify Clinical Response to csDMARD-Therapy and Predict Radiographic Progression Frances Humby1,* Myles Lewis1,* Nandhini Ramamoorthi2, Jason Hackney3, Michael Barnes1, Michele Bombardieri1, Francesca Setiadi2, Stephen Kelly1, Fabiola Bene1, Maria di Cicco1, Sudeh Riahi1, Vidalba Rocher-Ros1, Nora Ng1, Ilias Lazorou1, Rebecca E. Hands1, Desiree van der Heijde4, Robert Landewé5, Annette van der Helm-van Mil4, Alberto Cauli6, Iain B. McInnes7, Christopher D. Buckley8, Ernest Choy9, Peter Taylor10, Michael J. Townsend2 & Costantino Pitzalis1 1Centre for Experimental Medicine and Rheumatology, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, Charterhouse Square, London EC1M 6BQ, UK. Departments of 2Biomarker Discovery OMNI, 3Bioinformatics and Computational Biology, Genentech Research and Early Development, South San Francisco, California 94080 USA 4Department of Rheumatology, Leiden University Medical Center, The Netherlands 5Department of Clinical Immunology & Rheumatology, Amsterdam Rheumatology & Immunology Center, Amsterdam, The Netherlands 6Rheumatology Unit, Department of Medical Sciences, Policlinico of the University of Cagliari, Cagliari, Italy 7Institute of Infection, Immunity and Inflammation, University of Glasgow, Glasgow G12 8TA, UK 8Rheumatology Research Group, Institute of Inflammation and Ageing (IIA), University of Birmingham, Birmingham B15 2WB, UK 9Institute of -

Structural and Biochemical Changes Underlying a Keratoderma-Like Phenotype in Mice Lacking Suprabasal AP1 Transcription Factor Function
Citation: Cell Death and Disease (2015) 6, e1647; doi:10.1038/cddis.2015.21 OPEN & 2015 Macmillan Publishers Limited All rights reserved 2041-4889/15 www.nature.com/cddis Structural and biochemical changes underlying a keratoderma-like phenotype in mice lacking suprabasal AP1 transcription factor function EA Rorke*,1, G Adhikary2, CA Young2, RH Rice3, PM Elias4, D Crumrine4, J Meyer4, M Blumenberg5 and RL Eckert2,6,7,8 Epidermal keratinocyte differentiation on the body surface is a carefully choreographed process that leads to assembly of a barrier that is essential for life. Perturbation of keratinocyte differentiation leads to disease. Activator protein 1 (AP1) transcription factors are key controllers of this process. We have shown that inhibiting AP1 transcription factor activity in the suprabasal murine epidermis, by expression of dominant-negative c-jun (TAM67), produces a phenotype type that resembles human keratoderma. However, little is understood regarding the structural and molecular changes that drive this phenotype. In the present study we show that TAM67-positive epidermis displays altered cornified envelope, filaggrin-type keratohyalin granule, keratin filament, desmosome formation and lamellar body secretion leading to reduced barrier integrity. To understand the molecular changes underlying this process, we performed proteomic and RNA array analysis. Proteomic study of the corneocyte cross-linked proteome reveals a reduction in incorporation of cutaneous keratins, filaggrin, filaggrin2, late cornified envelope precursor proteins, hair keratins and hair keratin-associated proteins. This is coupled with increased incorporation of desmosome linker, small proline-rich, S100, transglutaminase and inflammation-associated proteins. Incorporation of most cutaneous keratins (Krt1, Krt5 and Krt10) is reduced, but incorporation of hyperproliferation-associated epidermal keratins (Krt6a, Krt6b and Krt16) is increased. -

CDH12 Cadherin 12, Type 2 N-Cadherin 2 RPL5 Ribosomal
5 6 6 5 . 4 2 1 1 1 2 4 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 A A A A A A A A A A A A A A A A A A A A C C C C C C C C C C C C C C C C C C C C R R R R R R R R R R R R R R R R R R R R B , B B B B B B B B B B B B B B B B B B B , 9 , , , , 4 , , 3 0 , , , , , , , , 6 2 , , 5 , 0 8 6 4 , 7 5 7 0 2 8 9 1 3 3 3 1 1 7 5 0 4 1 4 0 7 1 0 2 0 6 7 8 0 2 5 7 8 0 3 8 5 4 9 0 1 0 8 8 3 5 6 7 4 7 9 5 2 1 1 8 2 2 1 7 9 6 2 1 7 1 1 0 4 5 3 5 8 9 1 0 0 4 2 5 0 8 1 4 1 6 9 0 0 6 3 6 9 1 0 9 0 3 8 1 3 5 6 3 6 0 4 2 6 1 0 1 2 1 9 9 7 9 5 7 1 5 8 9 8 8 2 1 9 9 1 1 1 9 6 9 8 9 7 8 4 5 8 8 6 4 8 1 1 2 8 6 2 7 9 8 3 5 4 3 2 1 7 9 5 3 1 3 2 1 2 9 5 1 1 1 1 1 1 5 9 5 3 2 6 3 4 1 3 1 1 4 1 4 1 7 1 3 4 3 2 7 6 4 2 7 2 1 2 1 5 1 6 3 5 6 1 3 6 4 7 1 6 5 1 1 4 1 6 1 7 6 4 7 e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e e l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l l p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p p m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m m