Hypomesus Transpacificus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Development and Maintenance of Epidermal Stem Cells in Skin Adnexa
International Journal of Molecular Sciences Review Development and Maintenance of Epidermal Stem Cells in Skin Adnexa Jaroslav Mokry * and Rishikaysh Pisal Medical Faculty, Charles University, 500 03 Hradec Kralove, Czech Republic; [email protected] * Correspondence: [email protected] Received: 30 October 2020; Accepted: 18 December 2020; Published: 20 December 2020 Abstract: The skin surface is modified by numerous appendages. These structures arise from epithelial stem cells (SCs) through the induction of epidermal placodes as a result of local signalling interplay with mesenchymal cells based on the Wnt–(Dkk4)–Eda–Shh cascade. Slight modifications of the cascade, with the participation of antagonistic signalling, decide whether multipotent epidermal SCs develop in interfollicular epidermis, scales, hair/feather follicles, nails or skin glands. This review describes the roles of epidermal SCs in the development of skin adnexa and interfollicular epidermis, as well as their maintenance. Each skin structure arises from distinct pools of epidermal SCs that are harboured in specific but different niches that control SC behaviour. Such relationships explain differences in marker and gene expression patterns between particular SC subsets. The activity of well-compartmentalized epidermal SCs is orchestrated with that of other skin cells not only along the hair cycle but also in the course of skin regeneration following injury. This review highlights several membrane markers, cytoplasmic proteins and transcription factors associated with epidermal SCs. Keywords: stem cell; epidermal placode; skin adnexa; signalling; hair pigmentation; markers; keratins 1. Epidermal Stem Cells as Units of Development 1.1. Development of the Epidermis and Placode Formation The embryonic skin at very early stages of development is covered by a surface ectoderm that is a precursor to the epidermis and its multiple derivatives. -

Structural and Biochemical Changes Underlying a Keratoderma-Like Phenotype in Mice Lacking Suprabasal AP1 Transcription Factor Function
Citation: Cell Death and Disease (2015) 6, e1647; doi:10.1038/cddis.2015.21 OPEN & 2015 Macmillan Publishers Limited All rights reserved 2041-4889/15 www.nature.com/cddis Structural and biochemical changes underlying a keratoderma-like phenotype in mice lacking suprabasal AP1 transcription factor function EA Rorke*,1, G Adhikary2, CA Young2, RH Rice3, PM Elias4, D Crumrine4, J Meyer4, M Blumenberg5 and RL Eckert2,6,7,8 Epidermal keratinocyte differentiation on the body surface is a carefully choreographed process that leads to assembly of a barrier that is essential for life. Perturbation of keratinocyte differentiation leads to disease. Activator protein 1 (AP1) transcription factors are key controllers of this process. We have shown that inhibiting AP1 transcription factor activity in the suprabasal murine epidermis, by expression of dominant-negative c-jun (TAM67), produces a phenotype type that resembles human keratoderma. However, little is understood regarding the structural and molecular changes that drive this phenotype. In the present study we show that TAM67-positive epidermis displays altered cornified envelope, filaggrin-type keratohyalin granule, keratin filament, desmosome formation and lamellar body secretion leading to reduced barrier integrity. To understand the molecular changes underlying this process, we performed proteomic and RNA array analysis. Proteomic study of the corneocyte cross-linked proteome reveals a reduction in incorporation of cutaneous keratins, filaggrin, filaggrin2, late cornified envelope precursor proteins, hair keratins and hair keratin-associated proteins. This is coupled with increased incorporation of desmosome linker, small proline-rich, S100, transglutaminase and inflammation-associated proteins. Incorporation of most cutaneous keratins (Krt1, Krt5 and Krt10) is reduced, but incorporation of hyperproliferation-associated epidermal keratins (Krt6a, Krt6b and Krt16) is increased. -

Quantigene Flowrna Probe Sets Currently Available
QuantiGene FlowRNA Probe Sets Currently Available Accession No. Species Symbol Gene Name Catalog No. NM_003452 Human ZNF189 zinc finger protein 189 VA1-10009 NM_000057 Human BLM Bloom syndrome VA1-10010 NM_005269 Human GLI glioma-associated oncogene homolog (zinc finger protein) VA1-10011 NM_002614 Human PDZK1 PDZ domain containing 1 VA1-10015 NM_003225 Human TFF1 Trefoil factor 1 (breast cancer, estrogen-inducible sequence expressed in) VA1-10016 NM_002276 Human KRT19 keratin 19 VA1-10022 NM_002659 Human PLAUR plasminogen activator, urokinase receptor VA1-10025 NM_017669 Human ERCC6L excision repair cross-complementing rodent repair deficiency, complementation group 6-like VA1-10029 NM_017699 Human SIDT1 SID1 transmembrane family, member 1 VA1-10032 NM_000077 Human CDKN2A cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) VA1-10040 NM_003150 Human STAT3 signal transducer and activator of transcripton 3 (acute-phase response factor) VA1-10046 NM_004707 Human ATG12 ATG12 autophagy related 12 homolog (S. cerevisiae) VA1-10047 NM_000737 Human CGB chorionic gonadotropin, beta polypeptide VA1-10048 NM_001017420 Human ESCO2 establishment of cohesion 1 homolog 2 (S. cerevisiae) VA1-10050 NM_197978 Human HEMGN hemogen VA1-10051 NM_001738 Human CA1 Carbonic anhydrase I VA1-10052 NM_000184 Human HBG2 Hemoglobin, gamma G VA1-10053 NM_005330 Human HBE1 Hemoglobin, epsilon 1 VA1-10054 NR_003367 Human PVT1 Pvt1 oncogene homolog (mouse) VA1-10061 NM_000454 Human SOD1 Superoxide dismutase 1, soluble (amyotrophic lateral sclerosis 1 (adult)) -

140503 IPF Signatures Supplement Withfigs Thorax
Supplementary material for Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis Daryle J. DePianto1*, Sanjay Chandriani1⌘*, Alexander R. Abbas1, Guiquan Jia1, Elsa N. N’Diaye1, Patrick Caplazi1, Steven E. Kauder1, Sabyasachi Biswas1, Satyajit K. Karnik1#, Connie Ha1, Zora Modrusan1, Michael A. Matthay2, Jasleen Kukreja3, Harold R. Collard2, Jackson G. Egen1, Paul J. Wolters2§, and Joseph R. Arron1§ 1Genentech Research and Early Development, South San Francisco, CA 2Department of Medicine, University of California, San Francisco, CA 3Department of Surgery, University of California, San Francisco, CA ⌘Current address: Novartis Institutes for Biomedical Research, Emeryville, CA. #Current address: Gilead Sciences, Foster City, CA. *DJD and SC contributed equally to this manuscript §PJW and JRA co-directed this project Address correspondence to Paul J. Wolters, MD University of California, San Francisco Department of Medicine Box 0111 San Francisco, CA 94143-0111 [email protected] or Joseph R. Arron, MD, PhD Genentech, Inc. MS 231C 1 DNA Way South San Francisco, CA 94080 [email protected] 1 METHODS Human lung tissue samples Tissues were obtained at UCSF from clinical samples from IPF patients at the time of biopsy or lung transplantation. All patients were seen at UCSF and the diagnosis of IPF was established through multidisciplinary review of clinical, radiological, and pathological data according to criteria established by the consensus classification of the American Thoracic Society (ATS) and European Respiratory Society (ERS), Japanese Respiratory Society (JRS), and the Latin American Thoracic Association (ALAT) (ref. 5 in main text). Non-diseased normal lung tissues were procured from lungs not used by the Northern California Transplant Donor Network. -

Perlecan Expression Influences the Keratin 15-Positive Cell Population Fate in the Epidermis of Aging Skin
Perlecan expression influences the keratin 15-positive cell population fate in the epidermis of aging skin Morgan dos Santos, Anna Michopoulou, Valérie André-frei, Sophie Boulesteix, Christine Guicher, Guila Dayan, John Whitelock, Odile Damour, Patricia Rousselle To cite this version: Morgan dos Santos, Anna Michopoulou, Valérie André-frei, Sophie Boulesteix, Christine Guicher, et al.. Perlecan expression influences the keratin 15-positive cell population fate in the epidermis ofaging skin. Aging, Impact Journals, 2016, 8 (4), pp.751-768. 10.18632/aging.100928. hal-03041948 HAL Id: hal-03041948 https://hal.archives-ouvertes.fr/hal-03041948 Submitted on 5 Dec 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. www.impactaging.com AGING, April 2016, Vol 8 No 4 Research Paper Perlecan expression influences the keratin 15‐positive cell population fate in the epidermis of aging skin Morgan Dos Santos1,2,*, Anna Michopoulou1,*, Valérie André‐Frei2, Sophie Boulesteix1, Christine 1 1 3 1,4 1 Guicher , Guila Dayan , John Whitelock , Odile Damour , and Patricia -

The Correlation of Keratin Expression with In-Vitro Epithelial Cell Line Differentiation
The correlation of keratin expression with in-vitro epithelial cell line differentiation Deeqo Aden Thesis submitted to the University of London for Degree of Master of Philosophy (MPhil) Supervisors: Professor Ian. C. Mackenzie Professor Farida Fortune Centre for Clinical and Diagnostic Oral Science Barts and The London School of Medicine and Dentistry Queen Mary, University of London 2009 Contents Content pages ……………………………………………………………………......2 Abstract………………………………………………………………………….........6 Acknowledgements and Declaration……………………………………………...…7 List of Figures…………………………………………………………………………8 List of Tables………………………………………………………………………...12 Abbreviations….………………………………………………………………..…...14 Chapter 1: Literature review 16 1.1 Structure and function of the Oral Mucosa……………..…………….…..............17 1.2 Maintenance of the oral cavity...……………………………………….................20 1.2.1 Environmental Factors which damage the Oral Mucosa………. ….…………..21 1.3 Structure and function of the Oral Mucosa ………………...….……….………...21 1.3.1 Skin Barrier Formation………………………………………………….……...22 1.4 Comparison of Oral Mucosa and Skin…………………………………….……...24 1.5 Developmental and Experimental Models used in Oral mucosa and Skin...……..28 1.6 Keratinocytes…………………………………………………….….....................29 1.6.1 Desmosomes…………………………………………….…...............................29 1.6.2 Hemidesmosomes……………………………………….…...............................30 1.6.3 Tight Junctions………………………….……………….…...............................32 1.6.4 Gap Junctions………………………….……………….….................................32 -

Identification of Genes Involved in Regulatory Mechanism of Pigments in Broiler Chickens
Identification of genes involved in regulatory mechanism of pigments in broiler chickens T.M. Tarique1,2, S. Yang1, Z. Mohsina1, J. Qiu1, Z. Yan1, G. Chen1 and A. Chen1 1State Key Laboratory of Agro-Product Quality and Safety, Institute of Quality Standards and Testing Technology for Agro-Products, Chinese Academy of Agricultural Sciences, Beijing, China 2Department of Agricultural Sciences, Faculty of Sciences, Allama Iqbal Open University, Islamabad, Pakistan Corresponding authors: T.M. Tarique / S.M. Yang E-mail: [email protected] / [email protected] Genet. Mol. Res. 13 (3): 7201-7216 (2014) Received January 2, 2014 Accepted May 15, 2014 Published September 5, 2014 DOI http://dx.doi.org/10.4238/2014.September.5.6 ABSTRACT. Chicken is an important model organism that unites the evolutionary gap between mammals and other vertebrates and provide major source of protein from meat and eggs for all over the world population. However, specific genes underlying the regulatory mechanism of broiler pigmentation have not yet been determined. In order to better understand the genes involved in the mechanism of pigmentation in the muscle tissues of broilers, the Affymetrix microarray hybridization experiment platform was used to identify gene expression profiles at 7 weeks of age. Broilers fed canthaxanthin, natural lutein, and orangeII pigments (100 mg/kg) were used to explore gene expression profiles). Our data showed that the 7th week of age was a very important phase with regard to gene expression profiles. We identified a number of differentially expressed genes; in canthaxanthin, natural lutein, and orangeII, there were 54 (32 upregulated and 22 downregulated), 23 (15 upregulated and 8 downregulated), and 7 (5 upregulated and 2 downregulated) known genes, respectively. -

Product Data Sheet Purified Anti-Neurofilament H (NF-H)
Version: 2 Revision Date: 2015-10-26 Product Data Sheet Purified anti-Neurofilament H (NF-H) Catalog # / Size: 822601 / 50 µl Previously: Covance Catalog# PCK-592P Clone: Poly28226 Isotype: Chicken IgY Immunogen: This antibody was raised in chickens against purified bovine Neurofilament H. Reactivity: Human, Rat, Mouse, Cat, Chicken Preparation: The antibody was purified by affinity chromatography. Formulation: Phosphate-buffered solution, pH 7.2, containing 10 mM sodium azide. Concentration: Lot-specific (please contact technical support for concentration and total µg amount) Storage: The antibody solution should be stored undiluted between 2°C and 8°C. Please note the storage condition for this antibody has been changed Rat hippocampus stained with rabbit from -20°C to between 2°C and 8°C. You can also check your vial or your anti-GFAP (red, AF568) and chicken CoA to find the most accurate storage condition for this antibody. anti-Neurofilament H (green, AF488). Photo courtesy of Molecular Applications: Expressions. Applications: WB, IF Recommended Usage: Each lot of this antibody is quality control tested by Western blotting. The optimal working dilution should be determined for each specific assay condition. • WB: 1:1,000,000* • IF: 1:100,000 Application Notes: This antibody is effective in immunoblotting (WB) and immunofluorescence (IF). This antibody does not bind to protein A or protein G. * Predicted MW = 200-220 kD Description: Neurofilaments (NF) are ~10 nanometer intermediate filaments found in neurons. They are a major component of the neuronal cytoskeleton, and function primarily to provide structural support for the axon and regulate axon diameter. Neurofilaments belong to the same protein family as the intermediate filaments of other tissues such as keratins, which make the filaments expressed in epithelia. -
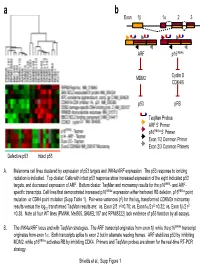
Taqman Probes ARF 5' Primer P16ink4a 5' Primer Exon 1/2
a b Exon 1β 1α 23 ARF p16INK4a MDM2 Cyclin D CDK4/6 p53 pRB TaqMan Probes ARF 5’ Primer p16INK4a 5’ Primer Exon 1/2 Common Primer Exon 2/3 Common Primers Defective p53 Intact p53 A. Melanoma cell lines clustered by expression of p53 targets and INK4a/ARF expression. The p53 response to ionizing radiation is indicated. Top cluster: Cells with intact p53 response show increased expression of the eight indicated p53 targets, and decreased expression of ARF. Bottom cluster: TaqMan and microarray results for the p16INK4- and ARF- specific transcripts. Cell lines that demonstrated increased p16INK4a expression either harbored RB deletion, p16INK4a point 2 mutation or CDK4 point mutation (Supp Table 1). Pair-wise variances (r ) for the log2 transformed CDKN2a microarray 2 2 2 results versus the log10 transformed TaqMan results are: vs. Exon 2/3 r =0.78; vs. Exon1α/2 r =0.32; vs. Exon 1β/2 r =0.38. Note all four WT lines (PMWK, Mel505, SKMEL187 and RPMI8322) lack evidence of p53 function by all assays. B. The INK4a/ARF locus and with TaqMan strategies. The ARF transcript originates from exon 1β while the p16INK4a transcript originates from exon 1α. Both transcripts splice to exon 2 but in alternate reading frames. ARF stabilizes p53 by inhibiting MDM2, while p16INK4a activates RB by inhibiting CDK4. Primers and TaqMan probes are shown for the real-time RT-PCR strategy. Shields et al., Supp Figure 1 SKMEL 28 U01 24h SKMEL WM2664 U01 48h WM2664 U01 24h 24 U01 48h SKMEL 24 U01 24h SKMEL 24 Untreated SKMEL 24 DMSO 48h SKMEL 24 DMSO 24h SKMEL WM2664 DMSO 24h WM2664 DMSO 48h WM2644 Untreated 28 Untreated SKMEL 28 DMSO 48h SKMEL 28 DMSO 24h SKMEL -3.00 -2.00 -1.00 0.00 1.00 2.00 3.00 relative to median expression Genes decreased by UO1 (863) HIF1A Hypoxia-inducible factor 1, alpha subunit NM_001530 RBBP8 Retinoblastoma binding protein 8 NM_002894 Homo sapiens, clone IMAGE:4337652, mRNA BC018676 EIF4EBP1 Eukaryotic translation initiation factor 4E binding protein EXOSC8 Exosome component 8 NM_181503 ENST00000321524 MCM7 MCM7 minichromosome maintenance deficient 7 (S. -

Transcriptome Profiling and Differential Gene Expression In
G C A T T A C G G C A T genes Article Transcriptome Profiling and Differential Gene Expression in Canine Microdissected Anagen and Telogen Hair Follicles and Interfollicular Epidermis Dominique J. Wiener 1,* ,Kátia R. Groch 1 , Magdalena A.T. Brunner 2,3, Tosso Leeb 2,3 , Vidhya Jagannathan 2 and Monika M. Welle 3,4 1 Department of Veterinary Pathobiology, College of Veterinary Medicine & Biomedical Science, Texas A&M University, College Station, TX 77843, USA; [email protected] 2 Institute of Genetics, Vetsuisse Faculty, University of Bern, 3012 Bern, Switzerland; [email protected] (M.A.T.B.); [email protected] (T.L.); [email protected] (V.J.) 3 Dermfocus, Vetsuisse Faculty, University Hospital of Bern, 3010 Bern, Switzerland; [email protected] 4 Institute of Animal Pathology, Vetsuisse Faculty, University of Bern, 3012 Bern, Switzerland * Correspondence: [email protected]; Tel.: +1-979-862-1568 Received: 30 June 2020; Accepted: 3 August 2020; Published: 4 August 2020 Abstract: The transcriptome profile and differential gene expression in telogen and late anagen microdissected hair follicles and the interfollicular epidermis of healthy dogs was investigated by using RNAseq. The genes with the highest expression levels in each group were identified and genes known from studies in other species to be associated with structure and function of hair follicles and epidermis were evaluated. Transcriptome profiling revealed that late anagen follicles expressed mainly keratins and telogen follicles expressed GSN and KRT15. The interfollicular epidermis expressed predominately genes encoding for proteins associated with differentiation. All sample groups express genes encoding for proteins involved in cellular growth and signal transduction. -

Characterization and Isolation of Stem Cell–Enriched Human Hair Follicle Bulge Cells
Characterization and isolation of stem cell–enriched human hair follicle bulge cells Manabu Ohyama, … , Mark C. Udey, Jonathan C. Vogel J Clin Invest. 2006;116(1):249-260. https://doi.org/10.1172/JCI26043. Research Article Dermatology The human hair follicle bulge is an important niche for keratinocyte stem cells (KSCs). Elucidation of human bulge cell biology could be facilitated by analysis of global gene expression profiles and identification of unique cell-surface markers. The lack of distinctive bulge morphology in human hair follicles has hampered studies of bulge cells and KSCs. In this study, we determined the distribution of label-retaining cells to define the human anagen bulge. Using navigated laser capture microdissection, bulge cells and outer root sheath cells from other follicle regions were obtained and analyzed with cDNA microarrays. Gene transcripts encoding inhibitors of WNT and activin/bone morphogenic protein signaling were overrepresented in the bulge, while genes responsible for cell proliferation were underrepresented, consistent with the existence of quiescent noncycling KSCs in anagen follicles. Positive markers for bulge cells included CD200, PHLDA1, follistatin, and frizzled homolog 1, while CD24, CD34, CD71, and CD146 were preferentially expressed by non- bulge keratinocytes. Importantly, CD200+ cells (CD200hiCD24loCD34loCD71loCD146lo) obtained from hair follicle suspensions demonstrated high colony-forming efficiency in clonogenic assays, indicating successful enrichment of living human bulge stem cells. The stem cell behavior of enriched bulge cells and their utility for gene therapy and hair regeneration will need to be assessed in in vivo assays. Find the latest version: https://jci.me/26043/pdf Related Commentary, page 19 Research article Characterization and isolation of stem cell– enriched human hair follicle bulge cells Manabu Ohyama,1 Atsushi Terunuma,1 Christine L. -

Comparative Proteomics Analysis of Primary Cutaneous Amyloidosis
3004 EXPERIMENTAL AND THERAPEUTIC MEDICINE 14: 3004-3012, 2017 Comparative proteomics analysis of primary cutaneous amyloidosis DAXING CAI1, YANG LI2, CHUNLEI ZHOU1, YULIN JIANG2, JIAN JIAO1 and LIN WU3 1Department of Dermatology, Qilu Hospital, Shandong University, Jinan, Shandong 250012; 2Beijing Protein Innovation Co., Ltd., Beijing 101318; 3Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, P.R. China Received August 16, 2016; Accepted May 25, 2017 DOI: 10.3892/etm.2017.4852 Abstract. Primary cutaneous amyloidosis (PCA) is a local- (adjusted P<0.001). KEGG analysis indicated that the upregu- ized skin disorder that is characterized by the abnormal lated proteins were significantly enriched in the signaling deposition of amyloid in the extracellular matrix (ECM) of pathways of cell communication, ECM receptor interaction the dermis. The pathogenesis of PCA is poorly understood. and focal adhesion (adjusted P<0.001). Furthermore, the The objective of the present study was to survey proteome upregulated proteins were enriched in the molecular func- changes in PCA lesions in order to gain insight into the tion of calcium ion binding, and the calcium binding proteins molecular basis and pathogenesis of PCA. Total protein from calmodulin-like protein 5, S100 calcium-binding protein A7 PCA lesions and normal skin tissue samples were extracted (S100A7)/fatty-acid binding protein and S100A8/A9 exhib- and analyzed using the isobaric tags for relative and abso- ited the highest levels of upregulation in PCA. This analysis lute quantitation technique. The function of differentially of differentially expressed proteins in PCA suggests that expressed proteins in PCA were analyzed by gene ontology increased focal adhesion, differentiation and wound healing is (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) associated with the pathogenesis of PCA.