Antibody Tools Immunohistochemistry
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

ZNF354C Is a Transcriptional Repressor That Inhibits Endothelial Angiogenic Sprouting James A
www.nature.com/scientificreports OPEN ZNF354C is a transcriptional repressor that inhibits endothelial angiogenic sprouting James A. Oo1,3, Barnabas Irmer1, Stefan Günther2, Timothy Warwick1, Katalin Pálf1, Judit Izquierdo Ponce1, Tom Teichmann1, Beatrice Pfüger‑Müller1,3, Ralf Gilsbach1,3, Ralf P. Brandes1,3 & Matthias S. Leisegang1,3* Zinc fnger proteins (ZNF) are a large group of transcription factors with diverse functions. We recently discovered that endothelial cells harbour a specifc mechanism to limit the action of ZNF354C, whose function in endothelial cells is unknown. Given that ZNF354C has so far only been studied in bone and tumour, its function was determined in endothelial cells. ZNF354C is expressed in vascular cells and localises to the nucleus and cytoplasm. Overexpression of ZNF354C in human endothelial cells results in a marked inhibition of endothelial sprouting. RNA‑sequencing of human microvascular endothelial cells with and without overexpression of ZNF354C revealed that the protein is a potent transcriptional repressor. ZNF354C contains an active KRAB domain which mediates this suppression as shown by mutagenesis analysis. ZNF354C interacts with dsDNA, TRIM28 and histones, as observed by proximity ligation and immunoprecipitation. Moreover, chromatin immunoprecipitation revealed that the ZNF binds to specifc endothelial‑relevant target‑gene promoters. ZNF354C suppresses these genes as shown by CRISPR/Cas knockout and RNAi. Inhibition of endothelial sprouting by ZNF354C is dependent on the amino acids DV and MLE of the KRAB domain. These results demonstrate that ZNF354C is a repressive transcription factor which acts through a KRAB domain to inhibit endothelial angiogenic sprouting. Te vascular system is controlled by numerous signaling pathways and growth factors which all contribute to the regulation of gene expression. -

Trastuzumab Modulates the Protein Cargo of Extracellular Vesicles Released by ERBB2+ Breast Can‐ Cer Cells
Supplementary Material: Trastuzumab Modulates the Protein Cargo of Extracellular Vesicles Released by ERBB2+ Breast Can‐ cer Cells Silvia Marconi, Sara Santamaria, Martina Bartolucci, Sara Stigliani, Cinzia Aiello, Maria Cristina Gagliani, Grazia Bellese, Andrea Petretto, Katia Cortese and Patrizio Castagnola Table S1. Antibodies used in the study. Antibody Catalog number Manufacturer Anti‐ALIX sc‐271975 Santa Cruz1 Anti‐CD9 PA5‐85955 Thermofisher Scientific2 Anti‐CD63 sc‐15363 Santa Cruz Anti‐ErbB2 (9G6) sc‐08 Santa Cruz Anti‐GAPDH 14C10 Cell signaling3 Anti‐HSP90 sc‐13119 Santa Cruz 1 Dallas, TX, USA; 2 Waltham, MA, USA; 3 Danvers, MA, USA. Table S2. Differentially regulated proteins by trastuzumab Tz treatment in extracellular vesicles EVs purified from SKBR‐ 3 cells with a statistically significant p‐value resulted from Studentʹs T‐test. The gene symbols coding for proteins downregulated by Tz (and hence upregulated in IgG treated cells) are highlighted in red while proteins upregulated by Tz are highlighted in blue. Official Gene Symbol 1 Gene product (Protein name) ACVR1B Activin A receptor type 1B ANO1 Anoctamin 1 ARFGEF2 ADP Ribosylation Factor Guanine Nucleotide Exchange Factor 2 BTN2A1 Butyrophilin subfamily 2 member A1 CIAPIN1 Cytokine Induced Apoptosis Inhibitor 1 CIT Citron Rho‐Interacting Serine/Threonine Kinase CPPED1 Calcineurin Like Phosphoesterase Domain Containing 1 DNAH7 Dynein Axonemal Heavy Chain 7 EIF3F Eukaryotic translation initiation factor 3 subunit F ESD Esterase D ESYT2 Extended Synaptotagmin 2 F2RL1 F2R Like Trypsin Receptor 1 RIPOR3 RIPOR Family Member 3 FZD6 Frizzled‐6 GAN Gigaxonin GTPBP2 GTP‐binding protein 2 GUCD1 Guanylyl Cyclase Domain Containing 1 HNRNPM Heterogeneous nuclear ribonucleoprotein M LMAN2 Lectin, Mannose Binding 2 LRRC8A Volume‐regulated anion channel subunit LRRC8A NOTCH4 Notch Receptor 4 NT5C2 5ʹ‐Nucleotidase, Cytosolic II PCID2 PCI domain‐containing 2 PDCD5 Programmed cell death 5 PHLDB3 Pleckstrin homology like domain family B member 3 PLAA Phospholipase A2 activating protein Membranes 2021, 11, 199. -

Genetic Basis of Simple and Complex Traits with Relevance to Avian Evolution
Genetic basis of simple and complex traits with relevance to avian evolution Małgorzata Anna Gazda Doctoral Program in Biodiversity, Genetics and Evolution D Faculdade de Ciências da Universidade do Porto 2019 Supervisor Miguel Jorge Pinto Carneiro, Auxiliary Researcher, CIBIO/InBIO, Laboratório Associado, Universidade do Porto Co-supervisor Ricardo Lopes, CIBIO/InBIO Leif Andersson, Uppsala University FCUP Genetic basis of avian traits Nota Previa Na elaboração desta tese, e nos termos do número 2 do Artigo 4º do Regulamento Geral dos Terceiros Ciclos de Estudos da Universidade do Porto e do Artigo 31º do D.L.74/2006, de 24 de Março, com a nova redação introduzida pelo D.L. 230/2009, de 14 de Setembro, foi efetuado o aproveitamento total de um conjunto coerente de trabalhos de investigação já publicados ou submetidos para publicação em revistas internacionais indexadas e com arbitragem científica, os quais integram alguns dos capítulos da presente tese. Tendo em conta que os referidos trabalhos foram realizados com a colaboração de outros autores, o candidato esclarece que, em todos eles, participou ativamente na sua conceção, na obtenção, análise e discussão de resultados, bem como na elaboração da sua forma publicada. Este trabalho foi apoiado pela Fundação para a Ciência e Tecnologia (FCT) através da atribuição de uma bolsa de doutoramento (PD/BD/114042/2015) no âmbito do programa doutoral em Biodiversidade, Genética e Evolução (BIODIV). 2 FCUP Genetic basis of avian traits Acknowledgements Firstly, I would like to thank to my all supervisors Miguel Carneiro, Ricardo Lopes and Leif Andersson, for the demanding task of supervising myself last four years. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Investigation of Candidate Genes and Mechanisms Underlying Obesity
Prashanth et al. BMC Endocrine Disorders (2021) 21:80 https://doi.org/10.1186/s12902-021-00718-5 RESEARCH ARTICLE Open Access Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules G. Prashanth1 , Basavaraj Vastrad2 , Anandkumar Tengli3 , Chanabasayya Vastrad4* and Iranna Kotturshetti5 Abstract Background: Obesity associated type 2 diabetes mellitus is a metabolic disorder ; however, the etiology of obesity associated type 2 diabetes mellitus remains largely unknown. There is an urgent need to further broaden the understanding of the molecular mechanism associated in obesity associated type 2 diabetes mellitus. Methods: To screen the differentially expressed genes (DEGs) that might play essential roles in obesity associated type 2 diabetes mellitus, the publicly available expression profiling by high throughput sequencing data (GSE143319) was downloaded and screened for DEGs. Then, Gene Ontology (GO) and REACTOME pathway enrichment analysis were performed. The protein - protein interaction network, miRNA - target genes regulatory network and TF-target gene regulatory network were constructed and analyzed for identification of hub and target genes. The hub genes were validated by receiver operating characteristic (ROC) curve analysis and RT- PCR analysis. Finally, a molecular docking study was performed on over expressed proteins to predict the target small drug molecules. Results: A total of 820 DEGs were identified between -

Α Are Regulated by Heat Shock Protein 90
The Levels of Retinoic Acid-Inducible Gene I Are Regulated by Heat Shock Protein 90- α Tomoh Matsumiya, Tadaatsu Imaizumi, Hidemi Yoshida, Kei Satoh, Matthew K. Topham and Diana M. Stafforini This information is current as of October 2, 2021. J Immunol 2009; 182:2717-2725; ; doi: 10.4049/jimmunol.0802933 http://www.jimmunol.org/content/182/5/2717 Downloaded from References This article cites 44 articles, 19 of which you can access for free at: http://www.jimmunol.org/content/182/5/2717.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on October 2, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2009 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Journal of Immunology The Levels of Retinoic Acid-Inducible Gene I Are Regulated by Heat Shock Protein 90-␣1 Tomoh Matsumiya,*‡ Tadaatsu Imaizumi,‡ Hidemi Yoshida,‡ Kei Satoh,‡ Matthew K. Topham,*† and Diana M. Stafforini2*† Retinoic acid-inducible gene I (RIG-I) is an intracellular pattern recognition receptor that plays important roles during innate immune responses to viral dsRNAs. -
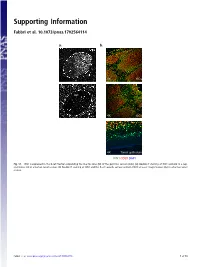
Supporting Information
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

Transcriptome Sequencing and Genome-Wide Association Analyses Reveal Lysosomal Function and Actin Cytoskeleton Remodeling in Schizophrenia and Bipolar Disorder
Molecular Psychiatry (2015) 20, 563–572 © 2015 Macmillan Publishers Limited All rights reserved 1359-4184/15 www.nature.com/mp ORIGINAL ARTICLE Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder Z Zhao1,6,JXu2,6, J Chen3,6, S Kim4, M Reimers3, S-A Bacanu3,HYu1, C Liu5, J Sun1, Q Wang1, P Jia1,FXu2, Y Zhang2, KS Kendler3, Z Peng2 and X Chen3 Schizophrenia (SCZ) and bipolar disorder (BPD) are severe mental disorders with high heritability. Clinicians have long noticed the similarities of clinic symptoms between these disorders. In recent years, accumulating evidence indicates some shared genetic liabilities. However, what is shared remains elusive. In this study, we conducted whole transcriptome analysis of post-mortem brain tissues (cingulate cortex) from SCZ, BPD and control subjects, and identified differentially expressed genes in these disorders. We found 105 and 153 genes differentially expressed in SCZ and BPD, respectively. By comparing the t-test scores, we found that many of the genes differentially expressed in SCZ and BPD are concordant in their expression level (q ⩽ 0.01, 53 genes; q ⩽ 0.05, 213 genes; q ⩽ 0.1, 885 genes). Using genome-wide association data from the Psychiatric Genomics Consortium, we found that these differentially and concordantly expressed genes were enriched in association signals for both SCZ (Po10 − 7) and BPD (P = 0.029). To our knowledge, this is the first time that a substantially large number of genes show concordant expression and association for both SCZ and BPD. Pathway analyses of these genes indicated that they are involved in the lysosome, Fc gamma receptor-mediated phagocytosis, regulation of actin cytoskeleton pathways, along with several cancer pathways. -

Chromatin Remodeling in Epstein-Barr Virus After Induction of the Lytic Phase: Molecular Characterization of the Role of Bzlf1 and Its Interactions
DISSERTATION ZUR ERLANGUNG DES DOKTORGRADES DER FAKULTÄT FÜR BIOLOGIE DER LUDWIG-MAXIMILIANS-UNIVERSITÄT MÜNCHEN CHROMATIN REMODELING IN EPSTEIN-BARR VIRUS AFTER INDUCTION OF THE LYTIC PHASE: MOLECULAR CHARACTERIZATION OF THE ROLE OF BZLF1 AND ITS INTERACTIONS MARISA SCHÄFFNER Dissertation eingereicht am 30. April 2015 Erstgutachter: Prof. Dr. Dirk Eick Zweitgutachter: Prof. Dr. Heinrich Leonhardt Tag der mündlichen Prüfung: 26.11.2015 ERKLÄRUNG Hiermit erkläre ich, dass die vorliegende Arbeit mit dem Titel „CHROMATIN REMODELING IN EPSTEIN-BARR VIRUS AFTER INDUCTION OF THE LYTIC PHASE: MOLECULAR CHARACTERIZATION OF THE ROLE OF BZLF1 AND ITS INTERACTIONS“ von mir selbstständig und ohne unerlaubte Hilfsmittel angefertigt wurde, und ich mich dabei nur der ausdrücklich bezeichneten Quellen und Hilfsmittel bedient habe. Die Arbeit wurde weder in der jetzigen noch in einer abgewandelten Form einer anderen Prüfungskommission vorgelegt. München, 30. April 2015 Marisa Schäffner CONTENT 1. Introduction....................................................................................................................... 7 1.1 The architecture of chromatin.......................................................................................... 7 1.1.1 Nucleosomes are histone octamers ........................................................................ 7 1.1.1.1 Nucleosome components.................................................................................. 7 1.1.1.2 Histone modifications and histone variants..................................................... -

Supplementary Table S1. Correlation Between the Mutant P53-Interacting Partners and PTTG3P, PTTG1 and PTTG2, Based on Data from Starbase V3.0 Database
Supplementary Table S1. Correlation between the mutant p53-interacting partners and PTTG3P, PTTG1 and PTTG2, based on data from StarBase v3.0 database. PTTG3P PTTG1 PTTG2 Gene ID Coefficient-R p-value Coefficient-R p-value Coefficient-R p-value NF-YA ENSG00000001167 −0.077 8.59e-2 −0.210 2.09e-6 −0.122 6.23e-3 NF-YB ENSG00000120837 0.176 7.12e-5 0.227 2.82e-7 0.094 3.59e-2 NF-YC ENSG00000066136 0.124 5.45e-3 0.124 5.40e-3 0.051 2.51e-1 Sp1 ENSG00000185591 −0.014 7.50e-1 −0.201 5.82e-6 −0.072 1.07e-1 Ets-1 ENSG00000134954 −0.096 3.14e-2 −0.257 4.83e-9 0.034 4.46e-1 VDR ENSG00000111424 −0.091 4.10e-2 −0.216 1.03e-6 0.014 7.48e-1 SREBP-2 ENSG00000198911 −0.064 1.53e-1 −0.147 9.27e-4 −0.073 1.01e-1 TopBP1 ENSG00000163781 0.067 1.36e-1 0.051 2.57e-1 −0.020 6.57e-1 Pin1 ENSG00000127445 0.250 1.40e-8 0.571 9.56e-45 0.187 2.52e-5 MRE11 ENSG00000020922 0.063 1.56e-1 −0.007 8.81e-1 −0.024 5.93e-1 PML ENSG00000140464 0.072 1.05e-1 0.217 9.36e-7 0.166 1.85e-4 p63 ENSG00000073282 −0.120 7.04e-3 −0.283 1.08e-10 −0.198 7.71e-6 p73 ENSG00000078900 0.104 2.03e-2 0.258 4.67e-9 0.097 3.02e-2 Supplementary Table S2.