Supplemental Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Educational Paper Ciliopathies
Eur J Pediatr (2012) 171:1285–1300 DOI 10.1007/s00431-011-1553-z REVIEW Educational paper Ciliopathies Carsten Bergmann Received: 11 June 2011 /Accepted: 3 August 2011 /Published online: 7 September 2011 # The Author(s) 2011. This article is published with open access at Springerlink.com Abstract Cilia are antenna-like organelles found on the (NPHP) . Ivemark syndrome . Meckel syndrome (MKS) . surface of most cells. They transduce molecular signals Joubert syndrome (JBTS) . Bardet–Biedl syndrome (BBS) . and facilitate interactions between cells and their Alstrom syndrome . Short-rib polydactyly syndromes . environment. Ciliary dysfunction has been shown to Jeune syndrome (ATD) . Ellis-van Crefeld syndrome (EVC) . underlie a broad range of overlapping, clinically and Sensenbrenner syndrome . Primary ciliary dyskinesia genetically heterogeneous phenotypes, collectively (Kartagener syndrome) . von Hippel-Lindau (VHL) . termed ciliopathies. Literally, all organs can be affected. Tuberous sclerosis (TSC) . Oligogenic inheritance . Modifier. Frequent cilia-related manifestations are (poly)cystic Mutational load kidney disease, retinal degeneration, situs inversus, cardiac defects, polydactyly, other skeletal abnormalities, and defects of the central and peripheral nervous Introduction system, occurring either isolated or as part of syn- dromes. Characterization of ciliopathies and the decisive Defective cellular organelles such as mitochondria, perox- role of primary cilia in signal transduction and cell isomes, and lysosomes are well-known -

A Heterozygous Variant in the Human Cardiac Mir-133 Gene, MIR133A2, Alters Mirna Duplex Processing and Strand Abundance
Ohanian et al. BMC Genetics 2013, 14:18 http://www.biomedcentral.com/1471-2156/14/18 RESEARCH ARTICLE Open Access A heterozygous variant in the human cardiac miR-133 gene, MIR133A2, alters miRNA duplex processing and strand abundance Monique Ohanian1†, David T Humphreys2,3†, Elizabeth Anderson4, Thomas Preiss5 and Diane Fatkin1,2,3,6* Abstract Background: MicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression. Sequential cleavage of miRNA precursors results in a ~22 nucleotide duplex of which one strand, the mature miRNA, is typically loaded into the RNA-induced silencing complex (RISC) while the passenger strand is degraded. Very little is known about how genetic variation might affect miRNA biogenesis and function. Results: We re-sequenced the MIR1-1, MIR1-2, MIR133A1, MIR133A2, and MIR133B genes, that encode the cardiac- enriched miRNAs, miR-1 and miR-133, in 120 individuals with familial atrial fibrillation and identified 10 variants, including a novel 79T > C MIR133A2 substitution. This variant lies within the duplex at the 30 end of the mature strand, miR-133a-3p, and is predicted to prevent base-pairing and weaken thermostability at this site, favoring incorporation of the passenger strand, miR-133a-5p, into RISC. Genomic DNA fragments containing miR-133a-2 precursor sequences with 79T and 79C alleles were transfected into HeLa cells. On Northern blotting the 79T allele showed strong expression of miR-133a-3p with weak expression of miR-133a-5p. In contrast, the 79C allele had no effect on miR-133a-3p but there was a significant increase (mean 3.6-fold) in miR-133a-5p levels. -

OR11H6 (NM 001004480) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC214860L3 OR11H6 (NM_001004480) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: OR11H6 (NM_001004480) Human Tagged ORF Clone Tag: Myc-DDK Symbol: OR11H6 Vector: pLenti-C-Myc-DDK-P2A-Puro (PS100092) E. coli Selection: Chloramphenicol (34 ug/mL) Cell Selection: Puromycin ORF Nucleotide The ORF insert of this clone is exactly the same as(RC214860). Sequence: Restriction Sites: SgfI-MluI Cloning Scheme: ACCN: NM_001004480 ORF Size: 990 bp This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 OR11H6 (NM_001004480) Human Tagged ORF Clone – RC214860L3 OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_001004480.1, NP_001004480.1 RefSeq Size: 993 bp RefSeq ORF: 993 bp Locus ID: 122748 UniProt ID: Q8NGC7, A0A126GVP4 Protein Families: Transmembrane Protein Pathways: Olfactory transduction MW: 36.6 kDa Gene Summary: Olfactory receptors interact with odorant molecules in the nose, to initiate a neuronal response that triggers the perception of a smell. -

Human Artificial Chromosome (Hac) Vector
Europäisches Patentamt *EP001559782A1* (19) European Patent Office Office européen des brevets (11) EP 1 559 782 A1 (12) EUROPEAN PATENT APPLICATION published in accordance with Art. 158(3) EPC (43) Date of publication: (51) Int Cl.7: C12N 15/09, C12N 1/15, 03.08.2005 Bulletin 2005/31 C12N 1/19, C12N 1/21, C12N 5/10, C12P 21/02 (21) Application number: 03751334.8 (86) International application number: (22) Date of filing: 03.10.2003 PCT/JP2003/012734 (87) International publication number: WO 2004/031385 (15.04.2004 Gazette 2004/16) (84) Designated Contracting States: • KATOH, Motonobu, Tottori University AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Yonago-shi, Tottori 683-8503 (JP) HU IE IT LI LU MC NL PT RO SE SI SK TR • TOMIZUKA, Kazuma, Designated Extension States: Kirin Beer Kabushiki Kaisha AL LT LV MK Takashi-shi, Gunma 370-1295 (JP) • KUROIWA, Yoshimi, (30) Priority: 04.10.2002 JP 2002292853 Kirin Beer Kabushiki Kaisha Takasaki-shi, Gunma 370-1295 (JP) (71) Applicant: KIRIN BEER KABUSHIKI KAISHA • KAKEDA, Minoru, Kirin Beer Kabushiki Kaisha Tokyo 104-8288 (JP) Takasaki-shi, Gunma 370-1295 (JP) (72) Inventors: (74) Representative: HOFFMANN - EITLE • OSHIMURA, Mitsuo, Tottori University Patent- und Rechtsanwälte Yonago-shi, Tottori 683-8503 (JP) Arabellastrasse 4 81925 München (DE) (54) HUMAN ARTIFICIAL CHROMOSOME (HAC) VECTOR (57) The present invention relates to a human arti- ing a cell which expresses foreign DNA. Furthermore, ficial chromosome (HAC) vector and a method for pro- the present invention relates to a method for producing ducing the same. -
Supporting Information
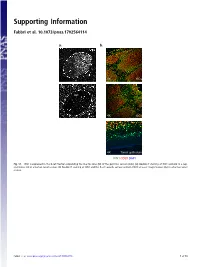
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

Ciliopathies Gene Panel
Ciliopathies Gene Panel Contact details Introduction Regional Genetics Service The ciliopathies are a heterogeneous group of conditions with considerable phenotypic overlap. Levels 4-6, Barclay House These inherited diseases are caused by defects in cilia; hair-like projections present on most 37 Queen Square cells, with roles in key human developmental processes via their motility and signalling functions. Ciliopathies are often lethal and multiple organ systems are affected. Ciliopathies are London, WC1N 3BH united in being genetically heterogeneous conditions and the different subtypes can share T +44 (0) 20 7762 6888 many clinical features, predominantly cystic kidney disease, but also retinal, respiratory, F +44 (0) 20 7813 8578 skeletal, hepatic and neurological defects in addition to metabolic defects, laterality defects and polydactyly. Their clinical variability can make ciliopathies hard to recognise, reflecting the ubiquity of cilia. Gene panels currently offer the best solution to tackling analysis of genetically Samples required heterogeneous conditions such as the ciliopathies. Ciliopathies affect approximately 1:2,000 5ml venous blood in plastic EDTA births. bottles (>1ml from neonates) Ciliopathies are generally inherited in an autosomal recessive manner, with some autosomal Prenatal testing must be arranged dominant and X-linked exceptions. in advance, through a Clinical Genetics department if possible. Referrals Amniotic fluid or CV samples Patients presenting with a ciliopathy; due to the phenotypic variability this could be a diverse set should be sent to Cytogenetics for of features. For guidance contact the laboratory or Dr Hannah Mitchison dissecting and culturing, with ([email protected]) / Prof Phil Beales ([email protected]) instructions to forward the sample to the Regional Molecular Genetics Referrals will be accepted from clinical geneticists and consultants in nephrology, metabolic, laboratory for analysis respiratory and retinal diseases. -

Supplementary Figure S4
18DCIS 18IDC Supplementary FigureS4 22DCIS 22IDC C D B A E (0.77) (0.78) 16DCIS 14DCIS 28DCIS 16IDC 28IDC (0.43) (0.49) 0 ADAMTS12 (p.E1469K) 14IDC ERBB2, LASP1,CDK12( CCNE1 ( NUTM2B SDHC,FCGR2B,PBX1,TPR( CD1D, B4GALT3, BCL9, FLG,NUP21OL,TPM3,TDRD10,RIT1,LMNA,PRCC,NTRK1 0 ADAMTS16 (p.E67K) (0.67) (0.89) (0.54) 0 ARHGEF38 (p.P179Hfs*29) 0 ATG9B (p.P823S) (0.68) (1.0) ARID5B, CCDC6 CCNE1, TSHZ3,CEP89 CREB3L2,TRIM24 BRAF, EGFR (7p11); 0 ABRACL (p.R35H) 0 CATSPER1 (p.P152H) 0 ADAMTS18 (p.Y799C) 19q12 0 CCDC88C (p.X1371_splice) (0) 0 ADRA1A (p.P327L) (10q22.3) 0 CCNF (p.D637N) −4 −2 −4 −2 0 AKAP4 (p.G454A) 0 CDYL (p.Y353Lfs*5) −4 −2 Log2 Ratio Log2 Ratio −4 −2 Log2 Ratio Log2 Ratio 0 2 4 0 2 4 0 ARID2 (p.R1068H) 0 COL27A1 (p.G646E) 0 2 4 0 2 4 2 EDRF1 (p.E521K) 0 ARPP21 (p.P791L) ) 0 DDX11 (p.E78K) 2 GPR101, p.A174V 0 ARPP21 (p.P791T) 0 DMGDH (p.W606C) 5 ANP32B, p.G237S 16IDC (Ploidy:2.01) 16DCIS (Ploidy:2.02) 14IDC (Ploidy:2.01) 14DCIS (Ploidy:2.9) -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 -3 -2 -1 Log Ratio Log Ratio Log Ratio Log Ratio 12DCIS 0 ASPM (p.S222T) Log Ratio Log Ratio 0 FMN2 (p.G941A) 20 1 2 3 2 0 1 2 3 2 ERBB3 (p.D297Y) 2 0 1 2 3 20 1 2 3 0 ATRX (p.L1276I) 20 1 2 3 2 0 1 2 3 0 GALNT18 (p.F92L) 2 MAPK4, p.H147Y 0 GALNTL6 (p.E236K) 5 C11orf1, p.Y53C (10q21.2); 0 ATRX (p.R1401W) PIK3CA, p.H1047R 28IDC (Ploidy:2.0) 28DCIS (Ploidy:2.0) 22IDC (Ploidy:3.7) 22DCIS (Ploidy:4.1) 18IDC (Ploidy:3.9) 18DCIS (Ploidy:2.3) 17q12 0 HCFC1 (p.S2025C) 2 LCMT1 (p.S34A) 0 ATXN7L2 (p.X453_splice) SPEN, p.P677Lfs*13 CBFB 1 2 3 4 5 6 7 8 9 10 11 -

Supplementary Methods
Heterogeneous Contribution of Microdeletions in the Development of Common Generalized and Focal epilepsies. SUPPLEMENTARY METHODS Epilepsy subtype extended description. Genetic Gereralized Epilepsy (GGE): Features unprovoked tonic and/or clonic seizures, originated inconsistently at some focal point within the brain that rapidly generalizes engaging bilateral distributed spikes and waves discharges on the electroencephalogram. This generalization can include cortical and sub cortical structures but not necessarily the entire cortex[1]. GGE is the most common group of epilepsies accounting for 20% of all cases[2]. It is characterized by an age-related onset and a strong familial aggregation and heritability which allows the assumption of a genetic cause. Although genetic associations have been identified, a broad spectrum of causes is acknowledged and remains largely unsolved [3]. Rolandic Epilepsy (RE): Commonly known also as Benign Epilepsy with Centrotemporal Spikes (BECTS), hallmarks early onset diagnosis (mean onset = 7 years old) with brief, focal hemifacial or oropharyngeal sensorimotor seizures alongside speech arrest and secondarily generalized tonic– clonic seizures, which mainly occur during sleep[4]. Rolandic epilepsy features a broad spectrum of less benign related syndromes called atypical Rolandic epilepsy (ARE), including benign partial epilepsy (ABPE), Landau–Kleffner syndrome(LKS) and epileptic encephalopathy with continuous spike-and-waves during sleep (CSWSS)[5]. Together they are the most common childhood epilepsy with a prevalence of 0.2–0.73/1000 (i.e. _1/2500)[6]. Adult Focal Epilepsy (AFE). Focal epilepsy is characterized by sporadic events of seizures originated within a specific brain region and restricted to one hemisphere. Although they can exhibit more than one network of wave discharges on the electroencephalogram, and different degrees of spreading, they feature a consistent site of origin. -

The Role of TGFβ Type III Receptor in Lung Cancer Cell Migration And
Western University Scholarship@Western Electronic Thesis and Dissertation Repository 6-26-2018 1:00 PM The role of TGFβ type III receptor in lung cancer cell migration and invasion Anthony Ziccarelli The University of Western Ontario Supervisor Di Guglielmo, John The University of Western Ontario Graduate Program in Physiology and Pharmacology A thesis submitted in partial fulfillment of the equirr ements for the degree in Master of Science © Anthony Ziccarelli 2018 Follow this and additional works at: https://ir.lib.uwo.ca/etd Recommended Citation Ziccarelli, Anthony, "The role of TGFβ type III receptor in lung cancer cell migration and invasion" (2018). Electronic Thesis and Dissertation Repository. 5454. https://ir.lib.uwo.ca/etd/5454 This Dissertation/Thesis is brought to you for free and open access by Scholarship@Western. It has been accepted for inclusion in Electronic Thesis and Dissertation Repository by an authorized administrator of Scholarship@Western. For more information, please contact [email protected]. i Abstract Metastasis is responsible for 90% of cancer-related deaths. An important early step in the metastatic process is epithelial-to-mesenchymal transition (EMT) of tumor cells. Stimulated by TGFβ signaling, cells that undergo EMT have increased migratory and invasive potential, resulting in metastasis and the development of tumors at a secondary site. The TGFβ type 3 receptor (TβR3) has been implicated in modulating TGFβ signaling, yet its functional outcomes remain unclear. My findings demonstrated that TβR3 silencing does not alter TGFβ-dependent Smad2 phosphorylation in neither H1299, not A549 non- small cell lung carcinoma cells but reduces Smad2 expression in H1299 cells. -

IBRAHIM ALDEAILEJ Phd.2013.Pdf
Bangor University DOCTOR OF PHILOSOPHY Identification and functional characterisation of germ line genes in human cancer cells Aldeailej, Ibrahim Award date: 2013 Awarding institution: Bangor University Link to publication General rights Copyright and moral rights for the publications made accessible in the public portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from the public portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain • You may freely distribute the URL identifying the publication in the public portal ? Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 09. Oct. 2021 Bangor University School of Biological Sciences Identification and functional characterisation of germ line genes in human cancer cells Ph.D. Thesis 2013 IBRAHIM ALDEAILEJ Declaration and Consent Details of the Work I hereby agree to deposit the following item in the digital repository maintained by Bangor University and/or in any other repository authorized for use by Bangor University. Author Name: IBRAHIM MOHAMMED ALDEAILEJ Title: Identification and functional characterisation of germ line genes in human cancer cells Supervisor/Department: Dr. Ramsay James McFarlane/ School of Biological Sciences Funding body (if any): kingdom of Saudi Arabia Government Qualification/Degree obtained: Ph.D. -

TULP1 Mutations Causing Early-Onset Retinal Degeneration: Preserved but Insensitive Macular Cones
Retina TULP1 Mutations Causing Early-Onset Retinal Degeneration: Preserved but Insensitive Macular Cones Samuel G. Jacobson,1 Artur V. Cideciyan,1 Wei Chieh Huang,1 Alexander Sumaroka,1 Alejandro J. Roman,1 Sharon B. Schwartz,1 Xunda Luo,1 Rebecca Sheplock,1 Joanna M. Dauber,1 Malgorzata Swider,1 and Edwin M. Stone2,3 1Scheie Eye Institute, Department of Ophthalmology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, United States 2Department of Ophthalmology, University of Iowa Carver College of Medicine, Iowa City, Iowa, United States 3Howard Hughes Medical Institute, Iowa City, Iowa, United States Correspondence: Samuel G. Jacob- PURPOSE. To investigate visual function and outer and inner retinal structure in the rare form of son, Scheie Eye Institute, University retinal degeneration (RD) caused by TULP1 (tubby-like protein 1) mutations. of Pennsylvania, 51 N. 39th Street, Philadelphia, PA 19104, USA; METHODS. Retinal degeneration patients with TULP1 mutations (n ¼ 5; age range, 5–36 years) [email protected]. were studied by kinetic and chromatic static perimetry, en face autofluorescence imaging, and spectral-domain optical coherence tomography (OCT) scans. Outer and inner retinal Submitted: April 10, 2014 Accepted: July 13, 2014 laminar thickness were measured and mapped across the central retina. Comparisons were made with results from patients with RD associated with four ciliopathy genotypes (MAK, Citation: Jacobson SG, Cideciyan AV, RPGR, BBS1, and USH2A). Huang WC, et al. TULP1 mutations causing early-onset retinal degenera- RESULTS. The TULP1-RD patients were severely affected already in the first decade of life and tion: preserved but insensitive macu- there was rapidly progressive visual loss. -

Supplementary Table 8. Cpcp PPI Network Details for Significantly Changed Proteins, As Identified in 3.2, Underlying Each of the Five Functional Domains
Supplementary Table 8. cPCP PPI network details for significantly changed proteins, as identified in 3.2, underlying each of the five functional domains. The network nodes represent each significant protein, followed by the list of interactors. Note that identifiers were converted to gene names to facilitate PPI database queries. Functional Domain Node Interactors Development and Park7 Rack1 differentiation Kcnma1 Atp6v1a Ywhae Ywhaz Pgls Hsd3b7 Development and Prdx6 Ncoa3 differentiation Pla2g4a Sufu Ncf2 Gstp1 Grin2b Ywhae Pgls Hsd3b7 Development and Atp1a2 Kcnma1 differentiation Vamp2 Development and Cntn1 Prnp differentiation Ywhaz Clstn1 Dlg4 App Ywhae Ywhab Development and Rac1 Pak1 differentiation Cdc42 Rhoa Dlg4 Ctnnb1 Mapk9 Mapk8 Pik3cb Sod1 Rrad Epb41l2 Nono Ltbp1 Evi5 Rbm39 Aplp2 Smurf2 Grin1 Grin2b Xiap Chn2 Cav1 Cybb Pgls Ywhae Development and Hbb-b1 Atp5b differentiation Hba Kcnma1 Got1 Aldoa Ywhaz Pgls Hsd3b4 Hsd3b7 Ywhae Development and Myh6 Mybpc3 differentiation Prkce Ywhae Development and Amph Capn2 differentiation Ap2a2 Dnm1 Dnm3 Dnm2 Atp6v1a Ywhab Development and Dnm3 Bin1 differentiation Amph Pacsin1 Grb2 Ywhae Bsn Development and Eef2 Ywhaz differentiation Rpgrip1l Atp6v1a Nphp1 Iqcb1 Ezh2 Ywhae Ywhab Pgls Hsd3b7 Hsd3b4 Development and Gnai1 Dlg4 differentiation Development and Gnao1 Dlg4 differentiation Vamp2 App Ywhae Ywhab Development and Psmd3 Rpgrip1l differentiation Psmd4 Hmga2 Development and Thy1 Syp differentiation Atp6v1a App Ywhae Ywhaz Ywhab Hsd3b7 Hsd3b4 Development and Tubb2a Ywhaz differentiation Nphp4