Transcriptional Co-Regulation of Micrornas and Protein-Coding Genes
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Analysis of Trans Esnps Infers Regulatory Network Architecture
Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Anat Kreimer All rights reserved ABSTRACT Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer eSNPs are genetic variants associated with transcript expression levels. The characteristics of such variants highlight their importance and present a unique opportunity for studying gene regulation. eSNPs affect most genes and their cell type specificity can shed light on different processes that are activated in each cell. They can identify functional variants by connecting SNPs that are implicated in disease to a molecular mechanism. Examining eSNPs that are associated with distal genes can provide insights regarding the inference of regulatory networks but also presents challenges due to the high statistical burden of multiple testing. Such association studies allow: simultaneous investigation of many gene expression phenotypes without assuming any prior knowledge and identification of unknown regulators of gene expression while uncovering directionality. This thesis will focus on such distal eSNPs to map regulatory interactions between different loci and expose the architecture of the regulatory network defined by such interactions. We develop novel computational approaches and apply them to genetics-genomics data in human. We go beyond pairwise interactions to define network motifs, including regulatory modules and bi-fan structures, showing them to be prevalent in real data and exposing distinct attributes of such arrangements. We project eSNP associations onto a protein-protein interaction network to expose topological properties of eSNPs and their targets and highlight different modes of distal regulation. -

Functional Roles of Bromodomain Proteins in Cancer
cancers Review Functional Roles of Bromodomain Proteins in Cancer Samuel P. Boyson 1,2, Cong Gao 3, Kathleen Quinn 2,3, Joseph Boyd 3, Hana Paculova 3 , Seth Frietze 3,4,* and Karen C. Glass 1,2,4,* 1 Department of Pharmaceutical Sciences, Albany College of Pharmacy and Health Sciences, Colchester, VT 05446, USA; [email protected] 2 Department of Pharmacology, Larner College of Medicine, University of Vermont, Burlington, VT 05405, USA; [email protected] 3 Department of Biomedical and Health Sciences, University of Vermont, Burlington, VT 05405, USA; [email protected] (C.G.); [email protected] (J.B.); [email protected] (H.P.) 4 University of Vermont Cancer Center, Burlington, VT 05405, USA * Correspondence: [email protected] (S.F.); [email protected] (K.C.G.) Simple Summary: This review provides an in depth analysis of the role of bromodomain-containing proteins in cancer development. As readers of acetylated lysine on nucleosomal histones, bromod- omain proteins are poised to activate gene expression, and often promote cancer progression. We examined changes in gene expression patterns that are observed in bromodomain-containing proteins and associated with specific cancer types. We also mapped the protein–protein interaction network for the human bromodomain-containing proteins, discuss the cellular roles of these epigenetic regu- lators as part of nine different functional groups, and identify bromodomain-specific mechanisms in cancer development. Lastly, we summarize emerging strategies to target bromodomain proteins in cancer therapy, including those that may be essential for overcoming resistance. Overall, this review provides a timely discussion of the different mechanisms of bromodomain-containing pro- Citation: Boyson, S.P.; Gao, C.; teins in cancer, and an updated assessment of their utility as a therapeutic target for a variety of Quinn, K.; Boyd, J.; Paculova, H.; cancer subtypes. -

Mitochondrial Dynamics During Spermatogenesis Grigor Varuzhanyan and David C
© 2020. Published by The Company of Biologists Ltd | Journal of Cell Science (2020) 133, jcs235937. doi:10.1242/jcs.235937 REVIEW SUBJECT COLLECTION: MITOCHONDRIA Mitochondrial dynamics during spermatogenesis Grigor Varuzhanyan and David C. Chan* ABSTRACT Mitophagy is an additional layer of quality control that utilizes Mitochondrial fusion and fission (mitochondrial dynamics) are autophagy (Mizushima, 2007; Mizushima et al., 1998; Tsukada and homeostatic processes that safeguard normal cellular function. This Ohsumi, 1993) to remove excessive mitochondria or damaged relationship is especially strong in tissues with constitutively high energy mitochondria that are beyond repair (Pickles et al., 2018). During demands, such as brain, heart and skeletal muscle. Less is known about mitophagy, microtubule-associated protein 1A/1B light chain 3 the role of mitochondrial dynamics in developmental systems that involve (collectively LC3; also known as MAP1LC3A, MAP1LC3B and changes in metabolic function. One such system is spermatogenesis. MAP1LC3C) is recruited to the autophagosomal membrane and binds The first mitochondrial dynamics gene, Fuzzy onions (Fzo), was to mitochondria that selectively express mitophagy receptors on their discovered in 1997 to mediate mitochondrial fusion during Drosophila outer membrane. This molecular recognition designates damaged spermatogenesis. In mammals, however, the role of mitochondrial fusion mitochondria as cargo for autophagosomes, and the resulting during spermatogenesis remained unknown for nearly two decades after mitophagosomes subsequently fuse with lysosomes for degradation discovery of Fzo. Mammalian spermatogenesis is one of the most and recycling of the engulfed organelles. Much of the molecular complex and lengthy differentiation processes in biology, transforming workings of mitophagy have been parsed in cultured cells, and spermatogonial stem cells into highly specialized sperm cells over a recently developed mouse models are enabling the analysis of 5-week period. -

A Correlation of Cytological and Genetical Crossing-Over in Zea Mays. PNAS 17:492–497
A CORRELATION OF CYTOLOGICAL AND GENETICAL CROSSING-OVER IN ZEA MAYS HARRIET B. CREIGHTON BARBARA MCCLINTOCK Botany Department Cornell University Ithaca, New York Creighton, H., and McClintock, B. 1931 A correlation of cytological and genetical crossing-over in Zea mays. PNAS 17:492–497. E S P Electronic Scholarly Publishing http://www.esp.org Electronic Scholarly Publishing Project Foundations Series –– Classical Genetics Series Editor: Robert J. Robbins The ESP Foundations of Classical Genetics project has received support from the ELSI component of the United States Department of Energy Human Genome Project. ESP also welcomes help from volunteers and collaborators, who recommend works for publication, provide access to original materials, and assist with technical and production work. If you are interested in volunteering, or are otherwise interested in the project, contact the series editor: [email protected]. Bibliographical Note This ESP edition, first electronically published in 2003 and subsequently revised in 2018, is a newly typeset, unabridged version, based on the 1931 edition published by The National Academy of Sciences. Unless explicitly noted, all footnotes and endnotes are as they appeared in the original work. Some of the graphics have been redone for this electronic version. Production Credits Scanning of originals: ESP staff OCRing of originals: ESP staff Typesetting: ESP staff Proofreading/Copyediting: ESP staff Graphics work: ESP staff Copyfitting/Final production: ESP staff © 2003, 2018 Electronic Scholarly Publishing Project http://www.esp.org This electronic edition is made freely available for educational or scholarly purposes, provided that this copyright notice is included. The manuscript may not be reprinted or redistributed for commercial purposes without permission. -

Quantitative Proteomics Methods for the Analysis of Histone Post-Translational Modifications
Université de Montréal Quantitative Proteomics Methods for the Analysis of Histone Post-translational Modifications par Nebiyu Ali Abshiru Département de Chimie Faculté des arts et des sciences Thèse présentée à la Faculté des études supérieures et postdoctorales en vue de l’obtention du grade de philosophiae doctor (Ph.D.) en chimie Septembre 2015 ©Nebiyu Ali Abshiru, 2015 i Résumé Les histones sont des protéines nucléaires hautement conservées chez les cellules des eucaryotes. Elles permettent d’organiser et de compacter l’ADN sous la forme de nucléosomes, ceux-ci representant les sous unités de base de la chromatine. Les histones peuvent être modifiées par de nombreuses modifications post-traductionnelles (PTMs) telles que l’acétylation, la méthylation et la phosphorylation. Ces modifications jouent un rôle essentiel dans la réplication de l’ADN, la transcription et l’assemblage de la chromatine. L’abondance de ces modifications peut varier de facon significative lors du developpement des maladies incluant plusieurs types de cancer. Par exemple, la perte totale de la triméthylation sur H4K20 ainsi que l’acétylation sur H4K16 sont des marqueurs tumoraux spécifiques a certains types de cancer chez l’humain. Par conséquent, l’étude de ces modifications et des événements determinant la dynamique des leurs changements d’abondance sont des atouts importants pour mieux comprendre les fonctions cellulaires et moléculaires lors du développement de la maladie. De manière générale, les modifications des histones sont étudiées par des approches biochimiques telles que les immuno-buvardage de type Western ou les méthodes d’immunoprécipitation de la chromatine (ChIP). Cependant, ces approches présentent plusieurs inconvénients telles que le manque de spécificité ou la disponibilité des anticorps, leur coût ou encore la difficulté de les produire et de les valider. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Protein Identities in Evs Isolated from U87-MG GBM Cells As Determined by NG LC-MS/MS
Protein identities in EVs isolated from U87-MG GBM cells as determined by NG LC-MS/MS. No. Accession Description Σ Coverage Σ# Proteins Σ# Unique Peptides Σ# Peptides Σ# PSMs # AAs MW [kDa] calc. pI 1 A8MS94 Putative golgin subfamily A member 2-like protein 5 OS=Homo sapiens PE=5 SV=2 - [GG2L5_HUMAN] 100 1 1 7 88 110 12,03704523 5,681152344 2 P60660 Myosin light polypeptide 6 OS=Homo sapiens GN=MYL6 PE=1 SV=2 - [MYL6_HUMAN] 100 3 5 17 173 151 16,91913397 4,652832031 3 Q6ZYL4 General transcription factor IIH subunit 5 OS=Homo sapiens GN=GTF2H5 PE=1 SV=1 - [TF2H5_HUMAN] 98,59 1 1 4 13 71 8,048185945 4,652832031 4 P60709 Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 - [ACTB_HUMAN] 97,6 5 5 35 917 375 41,70973209 5,478027344 5 P13489 Ribonuclease inhibitor OS=Homo sapiens GN=RNH1 PE=1 SV=2 - [RINI_HUMAN] 96,75 1 12 37 173 461 49,94108966 4,817871094 6 P09382 Galectin-1 OS=Homo sapiens GN=LGALS1 PE=1 SV=2 - [LEG1_HUMAN] 96,3 1 7 14 283 135 14,70620005 5,503417969 7 P60174 Triosephosphate isomerase OS=Homo sapiens GN=TPI1 PE=1 SV=3 - [TPIS_HUMAN] 95,1 3 16 25 375 286 30,77169764 5,922363281 8 P04406 Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 - [G3P_HUMAN] 94,63 2 13 31 509 335 36,03039959 8,455566406 9 Q15185 Prostaglandin E synthase 3 OS=Homo sapiens GN=PTGES3 PE=1 SV=1 - [TEBP_HUMAN] 93,13 1 5 12 74 160 18,68541938 4,538574219 10 P09417 Dihydropteridine reductase OS=Homo sapiens GN=QDPR PE=1 SV=2 - [DHPR_HUMAN] 93,03 1 1 17 69 244 25,77302971 7,371582031 11 P01911 HLA class II histocompatibility antigen, -

Introduction and Historical Perspective
Chapter 1 Introduction and Historical Perspective “ Nothing in biology makes sense except in the light of evolution. ” modified by the developmental history of the organism, Theodosius Dobzhansky its physiology – from cellular to systems levels – and by the social and physical environment. Finally, behaviors are shaped through evolutionary forces of natural selection OVERVIEW that optimize survival and reproduction ( Figure 1.1 ). Truly, the study of behavior provides us with a window through Behavioral genetics aims to understand the genetic which we can view much of biology. mechanisms that enable the nervous system to direct Understanding behaviors requires a multidisciplinary appropriate interactions between organisms and their perspective, with regulation of gene expression at its core. social and physical environments. Early scientific The emerging field of behavioral genetics is still taking explorations of animal behavior defined the fields shape and its boundaries are still being defined. Behavioral of experimental psychology and classical ethology. genetics has evolved through the merger of experimental Behavioral genetics has emerged as an interdisciplin- psychology and classical ethology with evolutionary biol- ary science at the interface of experimental psychology, ogy and genetics, and also incorporates aspects of neuro- classical ethology, genetics, and neuroscience. This science ( Figure 1.2 ). To gain a perspective on the current chapter provides a brief overview of the emergence of definition of this field, it is helpful -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -
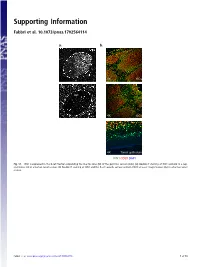
Supporting Information
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

Mir-17-92 Fine-Tunes MYC Expression and Function to Ensure
ARTICLE Received 31 Mar 2015 | Accepted 22 Sep 2015 | Published 10 Nov 2015 DOI: 10.1038/ncomms9725 OPEN miR-17-92 fine-tunes MYC expression and function to ensure optimal B cell lymphoma growth Marija Mihailovich1, Michael Bremang1, Valeria Spadotto1, Daniele Musiani1, Elena Vitale1, Gabriele Varano2,w, Federico Zambelli3, Francesco M. Mancuso1,w, David A. Cairns1,w, Giulio Pavesi3, Stefano Casola2 & Tiziana Bonaldi1 The synergism between c-MYC and miR-17-19b, a truncated version of the miR-17-92 cluster, is well-documented during tumor initiation. However, little is known about miR-17-19b function in established cancers. Here we investigate the role of miR-17-19b in c-MYC-driven lymphomas by integrating SILAC-based quantitative proteomics, transcriptomics and 30 untranslated region (UTR) analysis upon miR-17-19b overexpression. We identify over one hundred miR-17-19b targets, of which 40% are co-regulated by c-MYC. Downregulation of a new miR-17/20 target, checkpoint kinase 2 (Chek2), increases the recruitment of HuR to c- MYC transcripts, resulting in the inhibition of c-MYC translation and thus interfering with in vivo tumor growth. Hence, in established lymphomas, miR-17-19b fine-tunes c-MYC activity through a tight control of its function and expression, ultimately ensuring cancer cell homeostasis. Our data highlight the plasticity of miRNA function, reflecting changes in the mRNA landscape and 30 UTR shortening at different stages of tumorigenesis. 1 Department of Experimental Oncology, European Institute of Oncology, Via Adamello 16, Milan 20139, Italy. 2 Units of Genetics of B cells and lymphomas, IFOM, FIRC Institute of Molecular Oncology Foundation, Milan 20139, Italy. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase