Genomic Biomarkers Correlate with HLA-Identical Renal Transplant Tolerance
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Heterozygous Variant in the Human Cardiac Mir-133 Gene, MIR133A2, Alters Mirna Duplex Processing and Strand Abundance
Ohanian et al. BMC Genetics 2013, 14:18 http://www.biomedcentral.com/1471-2156/14/18 RESEARCH ARTICLE Open Access A heterozygous variant in the human cardiac miR-133 gene, MIR133A2, alters miRNA duplex processing and strand abundance Monique Ohanian1†, David T Humphreys2,3†, Elizabeth Anderson4, Thomas Preiss5 and Diane Fatkin1,2,3,6* Abstract Background: MicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression. Sequential cleavage of miRNA precursors results in a ~22 nucleotide duplex of which one strand, the mature miRNA, is typically loaded into the RNA-induced silencing complex (RISC) while the passenger strand is degraded. Very little is known about how genetic variation might affect miRNA biogenesis and function. Results: We re-sequenced the MIR1-1, MIR1-2, MIR133A1, MIR133A2, and MIR133B genes, that encode the cardiac- enriched miRNAs, miR-1 and miR-133, in 120 individuals with familial atrial fibrillation and identified 10 variants, including a novel 79T > C MIR133A2 substitution. This variant lies within the duplex at the 30 end of the mature strand, miR-133a-3p, and is predicted to prevent base-pairing and weaken thermostability at this site, favoring incorporation of the passenger strand, miR-133a-5p, into RISC. Genomic DNA fragments containing miR-133a-2 precursor sequences with 79T and 79C alleles were transfected into HeLa cells. On Northern blotting the 79T allele showed strong expression of miR-133a-3p with weak expression of miR-133a-5p. In contrast, the 79C allele had no effect on miR-133a-3p but there was a significant increase (mean 3.6-fold) in miR-133a-5p levels. -

Potassium Channels in Epilepsy
Downloaded from http://perspectivesinmedicine.cshlp.org/ on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Potassium Channels in Epilepsy Ru¨diger Ko¨hling and Jakob Wolfart Oscar Langendorff Institute of Physiology, University of Rostock, Rostock 18057, Germany Correspondence: [email protected] This review attempts to give a concise and up-to-date overview on the role of potassium channels in epilepsies. Their role can be defined from a genetic perspective, focusing on variants and de novo mutations identified in genetic studies or animal models with targeted, specific mutations in genes coding for a member of the large potassium channel family. In these genetic studies, a demonstrated functional link to hyperexcitability often remains elusive. However, their role can also be defined from a functional perspective, based on dy- namic, aggravating, or adaptive transcriptional and posttranslational alterations. In these cases, it often remains elusive whether the alteration is causal or merely incidental. With 80 potassium channel types, of which 10% are known to be associated with epilepsies (in humans) or a seizure phenotype (in animals), if genetically mutated, a comprehensive review is a challenging endeavor. This goal may seem all the more ambitious once the data on posttranslational alterations, found both in human tissue from epilepsy patients and in chronic or acute animal models, are included. We therefore summarize the literature, and expand only on key findings, particularly regarding functional alterations found in patient brain tissue and chronic animal models. INTRODUCTION TO POTASSIUM evolutionary appearance of voltage-gated so- CHANNELS dium (Nav)andcalcium (Cav)channels, Kchan- nels are further diversified in relation to their otassium (K) channels are related to epilepsy newer function, namely, keeping neuronal exci- Psyndromes on many different levels, ranging tation within limits (Anderson and Greenberg from direct control of neuronal excitability and 2001; Hille 2001). -

A Systematic Review on the Implications of O-Linked Glycan Branching and Truncating Enzymes on Cancer Progression and Metastasis
cells Review A Systematic Review on the Implications of O-linked Glycan Branching and Truncating Enzymes on Cancer Progression and Metastasis 1, 1, 1 1,2,3, Rohitesh Gupta y, Frank Leon y, Sanchita Rauth , Surinder K. Batra * and Moorthy P. Ponnusamy 1,2,* 1 Department of Biochemistry and Molecular Biology, University of Nebraska Medical Center, Omaha, NE 68105, USA; [email protected] (R.G.); [email protected] (F.L.); [email protected] (S.R.) 2 Fred and Pamela Buffett Cancer Center, Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, NE 681980-5900, USA 3 Department of Pathology and Microbiology, UNMC, Omaha, NE 68198-5900, USA * Correspondence: [email protected] (S.K.B.); [email protected] (M.P.P.); Tel.: +402-559-5455 (S.K.B.); +402-559-1170 (M.P.P.); Fax: +402-559-6650 (S.K.B. & M.P.P.) Equal contribution. y Received: 21 January 2020; Accepted: 12 February 2020; Published: 14 February 2020 Abstract: Glycosylation is the most commonly occurring post-translational modifications, and is believed to modify over 50% of all proteins. The process of glycan modification is directed by different glycosyltransferases, depending on the cell in which it is expressed. These small carbohydrate molecules consist of multiple glycan families that facilitate cell–cell interactions, protein interactions, and downstream signaling. An alteration of several types of O-glycan core structures have been implicated in multiple cancers, largely due to differential glycosyltransferase expression or activity. Consequently, aberrant O-linked glycosylation has been extensively demonstrated to affect biological function and protein integrity that directly result in cancer growth and progression of several diseases. -

Histatin-1 Attenuates LPS-Induced Inflammatory Signaling in RAW264
International Journal of Molecular Sciences Article Histatin-1 Attenuates LPS-Induced Inflammatory Signaling in RAW264.7 Macrophages Sang Min Lee 1 , Kyung-No Son 1, Dhara Shah 1, Marwan Ali 1, Arun Balasubramaniam 1, Deepak Shukla 1,2 and Vinay Kumar Aakalu 1,3,* 1 Department of Ophthalmology and Visual Sciences, University of Illinois at Chicago, Chicago, IL 60612, USA; [email protected] (S.M.L.); [email protected] (K.-N.S.); [email protected] (D.S.); [email protected] (M.A.); [email protected] (A.B.); [email protected] (D.S.) 2 Department of Microbiology and Immunology, University of Illinois at Chicago, Chicago, IL 60612, USA 3 Research and Surgical Services, Jesse Brown VA Medical Center, Chicago, IL 60612, USA * Correspondence: [email protected] Abstract: Macrophages play a critical role in the inflammatory response to environmental triggers, such as lipopolysaccharide (LPS). Inflammatory signaling through macrophages and the innate immune system are increasingly recognized as important contributors to multiple acute and chronic disease processes. Nitric oxide (NO) is a free radical that plays an important role in immune and inflammatory responses as an important intercellular messenger. In addition, NO has an important role in inflammatory responses in mucosal environments such as the ocular surface. Histatin peptides are well-established antimicrobial and wound healing agents. These peptides are important in multiple biological systems, playing roles in responses to the environment and immunomodulation. Citation: Lee, S.M.; Son, K.-N.; Shah, Given the importance of macrophages in responses to environmental triggers and pathogens, we D.; Ali, M.; Balasubramaniam, A.; Shukla, D.; Aakalu, V.K. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

A Novel Secretion and Online-Cleavage Strategy for Production of Cecropin a in Escherichia Coli
www.nature.com/scientificreports OPEN A novel secretion and online- cleavage strategy for production of cecropin A in Escherichia coli Received: 14 March 2017 Meng Wang 1, Minhua Huang1, Junjie Zhang1, Yi Ma1, Shan Li1 & Jufang Wang1,2 Accepted: 23 June 2017 Antimicrobial peptides, promising antibiotic candidates, are attracting increasing research attention. Published: xx xx xxxx Current methods for production of antimicrobial peptides are chemical synthesis, intracellular fusion expression, or direct separation and purifcation from natural sources. However, all these methods are costly, operation-complicated and low efciency. Here, we report a new strategy for extracellular secretion and online-cleavage of antimicrobial peptides on the surface of Escherichia coli, which is cost-efective, simple and does not require complex procedures like cell disruption and protein purifcation. Analysis by transmission electron microscopy and semi-denaturing detergent agarose gel electrophoresis indicated that fusion proteins contain cecropin A peptides can successfully be secreted and form extracellular amyloid aggregates at the surface of Escherichia coli on the basis of E. coli curli secretion system and amyloid characteristics of sup35NM. These amyloid aggregates can be easily collected by simple centrifugation and high-purity cecropin A peptide with the same antimicrobial activity as commercial peptide by chemical synthesis was released by efcient self-cleavage of Mxe GyrA intein. Here, we established a novel expression strategy for the production of antimicrobial peptides, which dramatically reduces the cost and simplifes purifcation procedures and gives new insights into producing antimicrobial and other commercially-viable peptides. Because of their potent, fast, long-lasting activity against a broad range of microorganisms and lack of bacterial resistance, antimicrobial peptides (AMPs) have received increasing attention1. -

OR11H6 (NM 001004480) Human Tagged ORF Clone Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for RC214860L3 OR11H6 (NM_001004480) Human Tagged ORF Clone Product data: Product Type: Expression Plasmids Product Name: OR11H6 (NM_001004480) Human Tagged ORF Clone Tag: Myc-DDK Symbol: OR11H6 Vector: pLenti-C-Myc-DDK-P2A-Puro (PS100092) E. coli Selection: Chloramphenicol (34 ug/mL) Cell Selection: Puromycin ORF Nucleotide The ORF insert of this clone is exactly the same as(RC214860). Sequence: Restriction Sites: SgfI-MluI Cloning Scheme: ACCN: NM_001004480 ORF Size: 990 bp This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 OR11H6 (NM_001004480) Human Tagged ORF Clone – RC214860L3 OTI Disclaimer: The molecular sequence of this clone aligns with the gene accession number as a point of reference only. However, individual transcript sequences of the same gene can differ through naturally occurring variations (e.g. polymorphisms), each with its own valid existence. This clone is substantially in agreement with the reference, but a complete review of all prevailing variants is recommended prior to use. More info OTI Annotation: This clone was engineered to express the complete ORF with an expression tag. Expression varies depending on the nature of the gene. RefSeq: NM_001004480.1, NP_001004480.1 RefSeq Size: 993 bp RefSeq ORF: 993 bp Locus ID: 122748 UniProt ID: Q8NGC7, A0A126GVP4 Protein Families: Transmembrane Protein Pathways: Olfactory transduction MW: 36.6 kDa Gene Summary: Olfactory receptors interact with odorant molecules in the nose, to initiate a neuronal response that triggers the perception of a smell. -

Human Artificial Chromosome (Hac) Vector
Europäisches Patentamt *EP001559782A1* (19) European Patent Office Office européen des brevets (11) EP 1 559 782 A1 (12) EUROPEAN PATENT APPLICATION published in accordance with Art. 158(3) EPC (43) Date of publication: (51) Int Cl.7: C12N 15/09, C12N 1/15, 03.08.2005 Bulletin 2005/31 C12N 1/19, C12N 1/21, C12N 5/10, C12P 21/02 (21) Application number: 03751334.8 (86) International application number: (22) Date of filing: 03.10.2003 PCT/JP2003/012734 (87) International publication number: WO 2004/031385 (15.04.2004 Gazette 2004/16) (84) Designated Contracting States: • KATOH, Motonobu, Tottori University AT BE BG CH CY CZ DE DK EE ES FI FR GB GR Yonago-shi, Tottori 683-8503 (JP) HU IE IT LI LU MC NL PT RO SE SI SK TR • TOMIZUKA, Kazuma, Designated Extension States: Kirin Beer Kabushiki Kaisha AL LT LV MK Takashi-shi, Gunma 370-1295 (JP) • KUROIWA, Yoshimi, (30) Priority: 04.10.2002 JP 2002292853 Kirin Beer Kabushiki Kaisha Takasaki-shi, Gunma 370-1295 (JP) (71) Applicant: KIRIN BEER KABUSHIKI KAISHA • KAKEDA, Minoru, Kirin Beer Kabushiki Kaisha Tokyo 104-8288 (JP) Takasaki-shi, Gunma 370-1295 (JP) (72) Inventors: (74) Representative: HOFFMANN - EITLE • OSHIMURA, Mitsuo, Tottori University Patent- und Rechtsanwälte Yonago-shi, Tottori 683-8503 (JP) Arabellastrasse 4 81925 München (DE) (54) HUMAN ARTIFICIAL CHROMOSOME (HAC) VECTOR (57) The present invention relates to a human arti- ing a cell which expresses foreign DNA. Furthermore, ficial chromosome (HAC) vector and a method for pro- the present invention relates to a method for producing ducing the same. -

Yeast Genome Gazetteer P35-65
gazetteer Metabolism 35 tRNA modification mitochondrial transport amino-acid metabolism other tRNA-transcription activities vesicular transport (Golgi network, etc.) nitrogen and sulphur metabolism mRNA synthesis peroxisomal transport nucleotide metabolism mRNA processing (splicing) vacuolar transport phosphate metabolism mRNA processing (5’-end, 3’-end processing extracellular transport carbohydrate metabolism and mRNA degradation) cellular import lipid, fatty-acid and sterol metabolism other mRNA-transcription activities other intracellular-transport activities biosynthesis of vitamins, cofactors and RNA transport prosthetic groups other transcription activities Cellular organization and biogenesis 54 ionic homeostasis organization and biogenesis of cell wall and Protein synthesis 48 plasma membrane Energy 40 ribosomal proteins organization and biogenesis of glycolysis translation (initiation,elongation and cytoskeleton gluconeogenesis termination) organization and biogenesis of endoplasmic pentose-phosphate pathway translational control reticulum and Golgi tricarboxylic-acid pathway tRNA synthetases organization and biogenesis of chromosome respiration other protein-synthesis activities structure fermentation mitochondrial organization and biogenesis metabolism of energy reserves (glycogen Protein destination 49 peroxisomal organization and biogenesis and trehalose) protein folding and stabilization endosomal organization and biogenesis other energy-generation activities protein targeting, sorting and translocation vacuolar and lysosomal -
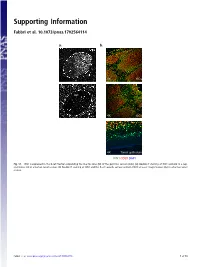
Supporting Information
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

Plenary and Platform Abstracts
American Society of Human Genetics 68th Annual Meeting PLENARY AND PLATFORM ABSTRACTS Abstract #'s Tuesday, October 16, 5:30-6:50 pm: 4. Featured Plenary Abstract Session I Hall C #1-#4 Wednesday, October 17, 9:00-10:00 am, Concurrent Platform Session A: 6. Variant Insights from Large Population Datasets Ballroom 20A #5-#8 7. GWAS in Combined Cancer Phenotypes Ballroom 20BC #9-#12 8. Genome-wide Epigenomics and Non-coding Variants Ballroom 20D #13-#16 9. Clonal Mosaicism in Cancer, Alzheimer's Disease, and Healthy Room 6A #17-#20 Tissue 10. Genetics of Behavioral Traits and Diseases Room 6B #21-#24 11. New Frontiers in Computational Genomics Room 6C #25-#28 12. Bone and Muscle: Identifying Causal Genes Room 6D #29-#32 13. Precision Medicine Initiatives: Outcomes and Lessons Learned Room 6E #33-#36 14. Environmental Exposures in Human Traits Room 6F #37-#40 Wednesday, October 17, 4:15-5:45 pm, Concurrent Platform Session B: 24. Variant Interpretation Practices and Resources Ballroom 20A #41-#46 25. Integrated Variant Analysis in Cancer Genomics Ballroom 20BC #47-#52 26. Gene Discovery and Functional Models of Neurological Disorders Ballroom 20D #53-#58 27. Whole Exome and Whole Genome Associations Room 6A #59-#64 28. Sequencing-based Diagnostics for Newborns and Infants Room 6B #65-#70 29. Omics Studies in Alzheimer's Disease Room 6C #71-#76 30. Cardiac, Valvular, and Vascular Disorders Room 6D #77-#82 31. Natural Selection and Human Phenotypes Room 6E #83-#88 32. Genetics of Cardiometabolic Traits Room 6F #89-#94 Wednesday, October 17, 6:00-7:00 pm, Concurrent Platform Session C: 33. -

High Expression of an Unknown Long Noncoding RNA RP11-290L1.3 from GDM Macrosomia and Its Effect on Preadipocyte Differentiation
ID: 20-0584 10 2 Y Lin, Y Zhand, L Xu et al. RP11 enhanced preadipocyte 10:2 191–204 differentiation RESEARCH High expression of an unknown long noncoding RNA RP11-290L1.3 from GDM macrosomia and its effect on preadipocyte differentiation Yu Lin1,*, Yingying Zhang1,*, Lei Xu2,*, Wei Long1, Chunjian Shan1, Hongjuan Ding1, Lianghui You1, Chun Zhao1 and Zhonghua Shi1 1State Key Laboratory of Reproductive Medicine, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, People’s Republic of China 2Maternal and Child Health Care Hospital of Dongchangfu District, Liaocheng, People’s Republic of China Correspondence should be addressed to C Zhao or Z Shi: [email protected] or [email protected] *(Y Lin, Y Zhang and L Xu contributed equally to this work) Abstract Aims: Gestational diabetes mellitus (GDM)-induced macrosomia is predominantly Key Words characterized by fat accumulation, which is closely related to adipocyte differentiation. f long noncoding RNA An unknown long noncoding RNA RP11-290L1.3, referred to as RP11, was identified to f GDM be dramatically upregulated in the umbilical cord blood of women with GDM-induced f preadipocyte macrosomia in our previous study. We conducted this study to identify the function of differentiation RP11 in GDM-induced macrosomia. f macrosomia Methods: The effects of RP11 gain- and loss-of-function on HPA-v (human preadipocytes- f fat accumulation visceral) adipogenesis were determined with lentivirus mediated cell transduction. The mRNA and protein expression levels of adipogenesis makers were evaluated by qPCR/Western blot. Then, we performed the microarray and pathway analysis to explore the possible mechanisms by which RP11 regulates adipogenesis.