UC Irvine UC Irvine Electronic Theses and Dissertations
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Prdm16 Is Required for the Maintenance of Neural Stem Cells in the Postnatal Forebrain and Their Differentiation Into Ependymal Cells
Downloaded from genesdev.cshlp.org on September 29, 2021 - Published by Cold Spring Harbor Laboratory Press Prdm16 is required for the maintenance of neural stem cells in the postnatal forebrain and their differentiation into ependymal cells Issei S. Shimada,1,2 Melih Acar,1,2,3 Rebecca J. Burgess,1,2 Zhiyu Zhao,1,2 and Sean J. Morrison1,2,4 1Children’s Research Institute, 2Department of Pediatrics, University of Texas Southwestern Medical Center, Dallas, Texas 75390, USA; 3Bahcesehir University, School of Medicine, Istanbul 34734, Turkey; 4Howard Hughes Medical Institute, University of Texas Southwestern Medical Center, Dallas, Texas 75390, USA We and others showed previously that PR domain-containing 16 (Prdm16) is a transcriptional regulator required for stem cell function in multiple fetal and neonatal tissues, including the nervous system. However, Prdm16 germline knockout mice died neonatally, preventing us from testing whether Prdm16 is also required for adult stem cell function. Here we demonstrate that Prdm16 is required for neural stem cell maintenance and neurogenesis in the adult lateral ventricle subventricular zone and dentate gyrus. We also discovered that Prdm16 is required for the formation of ciliated ependymal cells in the lateral ventricle. Conditional Prdm16 deletion during fetal development using Nestin-Cre prevented the formation of ependymal cells, disrupting cerebrospinal fluid flow and causing hy- drocephalus. Postnatal Prdm16 deletion using Nestin-CreERT2 did not cause hydrocephalus or prevent the forma- tion of ciliated ependymal cells but caused defects in their differentiation. Prdm16 was required in neural stem/ progenitor cells for the expression of Foxj1, a transcription factor that promotes ependymal cell differentiation. -

Downloaded from the National Database for Autism Research (NDAR)
International Journal of Molecular Sciences Article Phenotypic Subtyping and Re-Analysis of Existing Methylation Data from Autistic Probands in Simplex Families Reveal ASD Subtype-Associated Differentially Methylated Genes and Biological Functions Elizabeth C. Lee y and Valerie W. Hu * Department of Biochemistry and Molecular Medicine, The George Washington University, School of Medicine and Health Sciences, Washington, DC 20037, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-202-994-8431 Current address: W. Harry Feinstone Department of Molecular Microbiology and Immunology, y Johns Hopkins Bloomberg School of Public Health, Baltimore, MD 21205, USA. Received: 25 August 2020; Accepted: 17 September 2020; Published: 19 September 2020 Abstract: Autism spectrum disorder (ASD) describes a group of neurodevelopmental disorders with core deficits in social communication and manifestation of restricted, repetitive, and stereotyped behaviors. Despite the core symptomatology, ASD is extremely heterogeneous with respect to the severity of symptoms and behaviors. This heterogeneity presents an inherent challenge to all large-scale genome-wide omics analyses. In the present study, we address this heterogeneity by stratifying ASD probands from simplex families according to the severity of behavioral scores on the Autism Diagnostic Interview-Revised diagnostic instrument, followed by re-analysis of existing DNA methylation data from individuals in three ASD subphenotypes in comparison to that of their respective unaffected siblings. We demonstrate that subphenotyping of cases enables the identification of over 1.6 times the number of statistically significant differentially methylated regions (DMR) and DMR-associated genes (DAGs) between cases and controls, compared to that identified when all cases are combined. Our analyses also reveal ASD-related neurological functions and comorbidities that are enriched among DAGs in each phenotypic subgroup but not in the combined case group. -

Analysis of Trans Esnps Infers Regulatory Network Architecture
Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Anat Kreimer All rights reserved ABSTRACT Analysis of trans eSNPs infers regulatory network architecture Anat Kreimer eSNPs are genetic variants associated with transcript expression levels. The characteristics of such variants highlight their importance and present a unique opportunity for studying gene regulation. eSNPs affect most genes and their cell type specificity can shed light on different processes that are activated in each cell. They can identify functional variants by connecting SNPs that are implicated in disease to a molecular mechanism. Examining eSNPs that are associated with distal genes can provide insights regarding the inference of regulatory networks but also presents challenges due to the high statistical burden of multiple testing. Such association studies allow: simultaneous investigation of many gene expression phenotypes without assuming any prior knowledge and identification of unknown regulators of gene expression while uncovering directionality. This thesis will focus on such distal eSNPs to map regulatory interactions between different loci and expose the architecture of the regulatory network defined by such interactions. We develop novel computational approaches and apply them to genetics-genomics data in human. We go beyond pairwise interactions to define network motifs, including regulatory modules and bi-fan structures, showing them to be prevalent in real data and exposing distinct attributes of such arrangements. We project eSNP associations onto a protein-protein interaction network to expose topological properties of eSNPs and their targets and highlight different modes of distal regulation. -

Tomo-Seq Identifies SOX9 As a Key Regulator of Cardiac Fibrosis During Ischemic Injury
myocardial myocardial Eva van ◼ osis fibr SOX9 transcription ◼ PhD* PhD PhD PhD MSc, PhD naarden, MSc, PhD naarden, PhD* ventricular remodeling Correspondence to: Correspondence Rooij, MSc, PhD, Hubrecht Department of Institute, KNAW University Medical Cardiology, Uppsalalaan 8, Center Utrecht, The Netherlands. 3584CT Utrecht, E-mail [email protected] of Funding, see page 1408 Sources Key Words: ischemia ◼ © 2017 American Heart Association, Inc. *Drs. Lacraz and Junker contributed equally. Grégory P.A. Lacraz, MSc, P.A. Grégory MSc, Jan Philipp Junker, Monika M. Gladka, MSc, MSc Bas Molenaar, Scholman, MSc Koen T. MSc Marta Vigil-Garcia, BS Danielle Versteeg, BS Hesther de Ruiter, MSc, Vermunt, Marit W. MSc, Creyghton, Menno P. Manon M.H. Huibers, Nicolaas de Jonge, MD Alexander van Oude- Eva van Rooij, MSc, PhD 2017;136:1396–1409. DOI: 10.1161/CIRCULATIONAHA.117.027832 DOI: 2017;136:1396–1409. Circulation. blunted the cardiac fibrotic fibrotic blunted the cardiac Sox9 ). Subsequent correlation analysis allowed). Subsequent correlation Serca2 Editorial, see p 1410 , and Nppa Based on the exact local expression cues, tomo-seq can Based on the exact local expression Cardiac ischemic injury induces a pathological remodeling ischemic injury induces a pathological remodeling Cardiac , Although genome-wide transcriptome analysis on diseased tissues Tracing transcriptional differences with a high spatial resolution with a high spatial resolution transcriptional differences Tracing Col1a2 October 10, 2017 October 10, 1396 CONCLUSIONS: novel genes and key transcription factors involved in specific serve to reveal able to unveil the Using tomo-seq, we were remodeling. aspects of cardiac pointing fibrosis, of cardiac of SOX9 as a key regulator unknown relevance fibrosis. -

Contribution of the Potassium / Chloride Cotransporter KCC2 to Hippocampal Rhythmopathy Marie Goutierre
Contribution of the potassium / chloride cotransporter KCC2 to hippocampal rhythmopathy Marie Goutierre To cite this version: Marie Goutierre. Contribution of the potassium / chloride cotransporter KCC2 to hippocampal rhythmopathy. Neurobiology. Sorbonne Université, 2018. English. NNT : 2018SORUS600. tel- 02613734 HAL Id: tel-02613734 https://tel.archives-ouvertes.fr/tel-02613734 Submitted on 20 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Sorbonne Université Ecole doctorale n°158 - Cerveau Cognition Comportement - Institut du Fer à Moulin Equipe « Plasticité des réseaux corticaux et épilepsie » Contribution of the potassium / chloride cotransporter KCC2 to hippocampal rhythmopathy. Implication du transporteur potassium / chlore KCC2 dans la rythmopathie hippocampique. Par Marie GOUTIERRE Thèse de doctorat de Neurosciences Dirigée par Dr Jean-Christophe PONCER Présentée et soutenue publiquement le 28 juin 2018 Devant un jury composé de : Pr Ann LOHOF Présidente du jury Pr Claudio RIVERA Rapporteur Pr Andrew TREVELYAN Rapporteur Dr Lisa ROUX Examinateur Dr Corentin LE MAGUERESSE Examinateur Dr Jean-Christophe PONCER Directeur de thèse ABSTRACT In the CNS, synaptic release of the neurotransmitter GABA is responsible for fast inhibitory transmission. This is predominatly mediated by chloride flow through GABAA receptors. -

Potassium Channels in Epilepsy
Downloaded from http://perspectivesinmedicine.cshlp.org/ on September 28, 2021 - Published by Cold Spring Harbor Laboratory Press Potassium Channels in Epilepsy Ru¨diger Ko¨hling and Jakob Wolfart Oscar Langendorff Institute of Physiology, University of Rostock, Rostock 18057, Germany Correspondence: [email protected] This review attempts to give a concise and up-to-date overview on the role of potassium channels in epilepsies. Their role can be defined from a genetic perspective, focusing on variants and de novo mutations identified in genetic studies or animal models with targeted, specific mutations in genes coding for a member of the large potassium channel family. In these genetic studies, a demonstrated functional link to hyperexcitability often remains elusive. However, their role can also be defined from a functional perspective, based on dy- namic, aggravating, or adaptive transcriptional and posttranslational alterations. In these cases, it often remains elusive whether the alteration is causal or merely incidental. With 80 potassium channel types, of which 10% are known to be associated with epilepsies (in humans) or a seizure phenotype (in animals), if genetically mutated, a comprehensive review is a challenging endeavor. This goal may seem all the more ambitious once the data on posttranslational alterations, found both in human tissue from epilepsy patients and in chronic or acute animal models, are included. We therefore summarize the literature, and expand only on key findings, particularly regarding functional alterations found in patient brain tissue and chronic animal models. INTRODUCTION TO POTASSIUM evolutionary appearance of voltage-gated so- CHANNELS dium (Nav)andcalcium (Cav)channels, Kchan- nels are further diversified in relation to their otassium (K) channels are related to epilepsy newer function, namely, keeping neuronal exci- Psyndromes on many different levels, ranging tation within limits (Anderson and Greenberg from direct control of neuronal excitability and 2001; Hille 2001). -
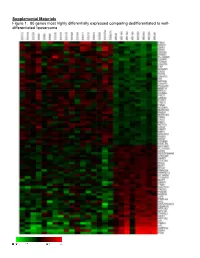
Supplemental Materials Figure 1. 80 Genes Most Highly Differentially Expressed Comparing Dedifferentiated to Well- Differentiated Liposarcoma
Supplemental Materials Figure 1. 80 genes most highly differentially expressed comparing dedifferentiated to well- differentiated liposarcoma Figure 2. Validation of microa CDC2, RACGAP1, FGFR-2, MAD2 and CITED1 200 CDK4 by Liposarcoma Subtype 16 0 12 0 80 Expression40 Level 0 Nl Fat rray data by quantitative RT-P Well Diff 16 0 p16 by Liposarcoma Subtype Dediff 12 0 80 Myxoid Expression40 Level Round Cell 0 12 0 MDM2 by Liposarcoma Subtype 10 0 Pleomorphic Nl Fat 80 60 40 Expression Level 20 RACGAP1 by LiposarcomaWell Diff Subtype 0 70 CR for genes CDK4, MDM2, p16, 60 Dediff 50 Nl Fat 40 30 Myxoid Expression20 Level Well Diff 10 50 0 Round Cell CDC2 by Liposarcoma Subtype Dediff 40 Nl Fat Pleomorphic 30 Myxoid 20 Expression Level Well Diff 10 MAD2L1 by Liposarcoma Subtype Round Cell 60 0 50 Dediff Pleomorphic 40 Nl Fat 30 Myxoid 20 Expression Level Well Diff 10 FGFR-2 by Liposarcoma Subtype 0 Round Cell 200 16 0 Dediff Nl Fat Pleomorphic 12 0 Myxoid 80 Expression Level Well Diff 40 Round Cell 0 Dediff Pleomorphic Nl Fat Myxoid Well Diff CITED1 by Liposarcoma Subtype Round Cell 10 0 80 Dediff Pleomorphic 60 40 Myxoid Expression Level 20 0 Round Cell Nl Fat Pleomorphic Well Diff Dediff Myxoid Round Cell Pleomorphic Table 1. 142 Gene classifier for liposarcoma subtype The genes used for each pair wise subtype comparison are grouped together. The flag column indicates which genes are unique to each subtype comparison. The values show the mean expression levels (actually the mean of the log expression levels was computed and than transformed back to absolute expression level). -

The Roles of Histone Deacetylase 5 and the Histone Methyltransferase Adaptor WDR5 in Myc Oncogenesis
The Roles of Histone Deacetylase 5 and the Histone Methyltransferase Adaptor WDR5 in Myc oncogenesis By Yuting Sun This thesis is submitted in fulfilment of the requirements for the degree of Doctor of Philosophy at the University of New South Wales Children’s Cancer Institute Australia for Medical Research School of Women’s and Children’s Health, Faculty of Medicine University of New South Wales Australia August 2014 PLEASE TYPE THE UNIVERSITY OF NEW SOUTH WALES Thesis/Dissertation Sheet Surname or Family name: Sun First name: Yuting Other name/s: Abbreviation for degree as given in the University calendar: PhD School : School of·Women's and Children's Health Faculty: Faculty of Medicine Title: The Roles of Histone Deacetylase 5 and the Histone Methyltransferase Adaptor WDR5 in Myc oncogenesis. Abstract 350 words maximum: (PLEASE TYPE) N-Myc Induces neuroblastoma by regulating the expression of target genes and proteins, and N-Myc protein is degraded by Fbxw7 and NEDD4 and stabilized by Aurora A. The class lla histone deacetylase HDAC5 suppresses gene transcription, and blocks myoblast and leukaemia cell differentiation. While histone H3 lysine 4 (H3K4) trimethylation at target gene promoters is a pre-requisite for Myc· induced transcriptional activation, WDRS, as a histone H3K4 methyltransferase presenter, is required for H3K4 methylation and transcriptional activation mediated by a histone H3K4 methyltransferase complex. Here, I investigated the roles of HDAC5 and WDR5 in N-Myc overexpressing neuroblastoma. I have found that N-Myc upregulates HDAC5 protein expression, and that HDAC5 represses NEDD4 gene expression, increases Aurora A gene expression and consequently upregulates N-Myc protein expression in neuroblastoma cells. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Ubiquitylome Profiling of Parkin-Null Brain Reveals Dysregulation Of
Neurobiology of Disease 127 (2019) 114–130 Contents lists available at ScienceDirect Neurobiology of Disease journal homepage: www.elsevier.com/locate/ynbdi Ubiquitylome profiling of Parkin-null brain reveals dysregulation of calcium T homeostasis factors ATP1A2, Hippocalcin and GNA11, reflected by altered firing of noradrenergic neurons Key J.a,1, Mueller A.K.b,1, Gispert S.a, Matschke L.b, Wittig I.c, Corti O.d,e,f,g, Münch C.h, ⁎ ⁎ Decher N.b, , Auburger G.a, a Exp. Neurology, Goethe University Medical School, 60590 Frankfurt am Main, Germany b Institute for Physiology and Pathophysiology, Vegetative Physiology and Marburg Center for Mind, Brain and Behavior - MCMBB; Clinic for Neurology, Philipps-University Marburg, 35037 Marburg, Germany c Functional Proteomics, SFB 815 Core Unit, Goethe University Medical School, 60590 Frankfurt am Main, Germany d Institut du Cerveau et de la Moelle épinière, ICM, Paris, F-75013, France e Inserm, U1127, Paris, F-75013, France f CNRS, UMR 7225, Paris, F-75013, France g Sorbonne Universités, Paris, F-75013, France h Institute of Biochemistry II, Goethe University Medical School, 60590 Frankfurt am Main, Germany ARTICLE INFO ABSTRACT Keywords: Parkinson's disease (PD) is the second most frequent neurodegenerative disorder in the old population. Among Parkinson's disease its monogenic variants, a frequent cause is a mutation in the Parkin gene (Prkn). Deficient function of Parkin Mitochondria triggers ubiquitous mitochondrial dysfunction and inflammation in the brain, but it remains unclear howse- Parkin lective neural circuits become vulnerable and finally undergo atrophy. Ubiquitin We attempted to go beyond previous work, mostly done in peripheral tumor cells, which identified protein Calcium targets of Parkin activity, an ubiquitin E3 ligase. -
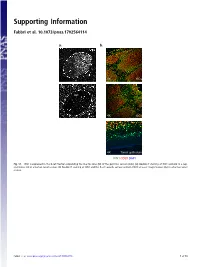
Supporting Information
Supporting Information Fabbri et al. 10.1073/pnas.1702564114 a. b. GC 4X GCs 20x GC 4X GCs 4X Tonsil epithelium ICN1 CD20 DAPI Fig. S1. ICN1 is expressed in the B-cell fraction populating the mantle zone (M) of the germinal centers (GCs). (A) Double IF staining of ICN1 and AID in a rep- resentative GC in a human tonsil section. (B) Double IF staining of ICN1 and the B-cell–specific surface antigen CD20 at lower magnification (4×) in a human tonsil section. Fabbri et al. www.pnas.org/cgi/content/short/1702564114 1of10 Fig. S2. ICN1 expression analysis in a panel of primary CLL cases and PBMC. (A) IB analysis of ICN1 and control β-actin in a panel of 124 CLL PB primary CLL cases, (B) in primary NOTCH1–wild-type CLL cells treated with the γ-secretase inhibitor Compound E (CpE, 500 nM, 8 h) or control DMSO, and (C) in PBMC protein extracts and representative primary CLL cases expressing ICN1. Samples are color-coded based on the NOTCH1 mutational status [red, clonal NOTCH1 PEST-truncating events; orange, subclonal NOTCH1 PEST-truncating events; blue, RAG-mediated NOTCH1 translocation (83); and black, NOTCH1–wild-type]. Samples in gray were excluded from the analysis because of low quality of the protein lysate, low viability, or low leukemic representation. Color-coded arrows indicate cases subjected to RNA-Seq analysis: dark red denotes NOTCH1-mutated cases expressing ICN1; blue, NOTCH1–wild-type cases expressing ICN1; and green, ICN1− NOTCH1–wild-type cases. Abbreviations: MO+DL1, MO1043 cells cocultured on OP9-DL1 cells (54); s.e., short exposure; l.e., long exposure. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like,