Wounding and Insect Feeding Trigger Two Independent MAPK Pathways with Distinct 25 Regulation and Kinetics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Significant Shortest Paths for the Detection of Putative Disease Modules
bioRxiv preprint doi: https://doi.org/10.1101/2020.04.01.019844; this version posted April 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. SIGNIFICANT SHORTEST PATHS FOR THE DETECTION OF PUTATIVE DISEASE MODULES Daniele Pepe1 1Department of Oncology, KU Leuven, LKI–Leuven Cancer Institute, Leuven, Belgium Email address: DP: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.04.01.019844; this version posted April 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Keywords Structural equation modeling, significant shortest paths, pathway analysis, disease modules. Abstract Background The characterization of diseases in terms of perturbated gene modules was recently introduced for the analysis of gene expression data. Some approaches were proposed in literature, but many times they are inductive approaches. This means that starting directly from data, they try to infer key gene networks potentially associated to the biological phenomenon studied. However they ignore the biological information already available to characterize the gene modules. Here we propose the detection of perturbed gene modules using the combination of data driven and hypothesis-driven approaches relying on biological metabolic pathways and significant shortest paths tested by structural equation modeling. -

Advancing a Clinically Relevant Perspective of the Clonal Nature of Cancer
Advancing a clinically relevant perspective of the clonal nature of cancer Christian Ruiza,b, Elizabeth Lenkiewicza, Lisa Eversa, Tara Holleya, Alex Robesona, Jeffrey Kieferc, Michael J. Demeurea,d, Michael A. Hollingsworthe, Michael Shenf, Donna Prunkardf, Peter S. Rabinovitchf, Tobias Zellwegerg, Spyro Moussesc, Jeffrey M. Trenta,h, John D. Carpteni, Lukas Bubendorfb, Daniel Von Hoffa,d, and Michael T. Barretta,1 aClinical Translational Research Division, Translational Genomics Research Institute, Scottsdale, AZ 85259; bInstitute for Pathology, University Hospital Basel, University of Basel, 4031 Basel, Switzerland; cGenetic Basis of Human Disease, Translational Genomics Research Institute, Phoenix, AZ 85004; dVirginia G. Piper Cancer Center, Scottsdale Healthcare, Scottsdale, AZ 85258; eEppley Institute for Research in Cancer and Allied Diseases, Nebraska Medical Center, Omaha, NE 68198; fDepartment of Pathology, University of Washington, Seattle, WA 98105; gDivision of Urology, St. Claraspital and University of Basel, 4058 Basel, Switzerland; hVan Andel Research Institute, Grand Rapids, MI 49503; and iIntegrated Cancer Genomics Division, Translational Genomics Research Institute, Phoenix, AZ 85004 Edited* by George F. Vande Woude, Van Andel Research Institute, Grand Rapids, MI, and approved June 10, 2011 (received for review March 11, 2011) Cancers frequently arise as a result of an acquired genomic insta- on the basis of morphology alone (8). Thus, the application of bility and the subsequent clonal evolution of neoplastic cells with purification methods such as laser capture microdissection does variable patterns of genetic aberrations. Thus, the presence and not resolve the complexities of many samples. A second approach behaviors of distinct clonal populations in each patient’s tumor is to passage tumor biopsies in tissue culture or in xenografts (4, 9– may underlie multiple clinical phenotypes in cancers. -

Interactions Between the Parasite Philasterides Dicentrarchi and the Immune System of the Turbot Scophthalmus Maximus.A Transcriptomic Analysis
biology Article Interactions between the Parasite Philasterides dicentrarchi and the Immune System of the Turbot Scophthalmus maximus.A Transcriptomic Analysis Alejandra Valle 1 , José Manuel Leiro 2 , Patricia Pereiro 3 , Antonio Figueras 3 , Beatriz Novoa 3, Ron P. H. Dirks 4 and Jesús Lamas 1,* 1 Department of Fundamental Biology, Institute of Aquaculture, Campus Vida, University of Santiago de Compostela, 15782 Santiago de Compostela, Spain; [email protected] 2 Department of Microbiology and Parasitology, Laboratory of Parasitology, Institute of Research on Chemical and Biological Analysis, Campus Vida, University of Santiago de Compostela, 15782 Santiago de Compostela, Spain; [email protected] 3 Institute of Marine Research, Consejo Superior de Investigaciones Científicas-CSIC, 36208 Vigo, Spain; [email protected] (P.P.); antoniofi[email protected] (A.F.); [email protected] (B.N.) 4 Future Genomics Technologies, Leiden BioScience Park, 2333 BE Leiden, The Netherlands; [email protected] * Correspondence: [email protected]; Tel.: +34-88-181-6951; Fax: +34-88-159-6904 Received: 4 September 2020; Accepted: 14 October 2020; Published: 15 October 2020 Simple Summary: Philasterides dicentrarchi is a free-living ciliate that causes high mortality in marine cultured fish, particularly flatfish, and in fish kept in aquaria. At present, there is still no clear picture of what makes this ciliate a fish pathogen and what makes fish resistant to this ciliate. In the present study, we used transcriptomic techniques to evaluate the interactions between P. dicentrarchi and turbot leucocytes during the early stages of infection. The findings enabled us to identify some parasite genes/proteins that may be involved in virulence and host resistance, some of which may be good candidates for inclusion in fish vaccines. -

Modulation of NF-Κb Signalling by Microbial Pathogens
REVIEWS Modulation of NF‑κB signalling by microbial pathogens Masmudur M. Rahman and Grant McFadden Abstract | The nuclear factor-κB (NF‑κB) family of transcription factors plays a central part in the host response to infection by microbial pathogens, by orchestrating the innate and acquired host immune responses. The NF‑κB proteins are activated by diverse signalling pathways that originate from many different cellular receptors and sensors. Many successful pathogens have acquired sophisticated mechanisms to regulate the NF‑κB signalling pathways by deploying subversive proteins or hijacking the host signalling molecules. Here, we describe the mechanisms by which viruses and bacteria micromanage the host NF‑κB signalling circuitry to favour the continued survival of the pathogen. The nuclear factor-κB (NF-κB) family of transcription Signalling targets upstream of NF‑κB factors regulates the expression of hundreds of genes that NF-κB proteins are tightly regulated in both the cyto- are associated with diverse cellular processes, such as pro- plasm and the nucleus6. Under normal physiological liferation, differentiation and death, as well as innate and conditions, NF‑κB complexes remain inactive in the adaptive immune responses. The mammalian NF‑κB cytoplasm through a direct interaction with proteins proteins are members of the Rel domain-containing pro- of the inhibitor of NF-κB (IκB) family, including IκBα, tein family: RELA (also known as p65), RELB, c‑REL, IκBβ and IκBε (also known as NF-κBIα, NF-κBIβ and the NF-κB p105 subunit (also known as NF‑κB1; which NF-κBIε, respectively); IκB proteins mask the nuclear is cleaved into the p50 subunit) and the NF-κB p100 localization domains in the NF‑κB complex, thus subunit (also known as NF‑κB2; which is cleaved into retaining the transcription complex in the cytoplasm. -

PRODUCT SPECIFICATION Anti-MAP3K10 Product Datasheet
Anti-MAP3K10 Product Datasheet Polyclonal Antibody PRODUCT SPECIFICATION Product Name Anti-MAP3K10 Product Number HPA007039 Gene Description mitogen-activated protein kinase kinase kinase 10 Clonality Polyclonal Isotype IgG Host Rabbit Antigen Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: LPSGFEHKITVQASPTLDKRKGSDGASPPASPSIIPRLRAIRLTPVDCGG SSSGSSSGGSGTWSRGGPPKKEELVGGKKKGRTWGPSSTLQKERVGGEER LKGLGEGSKQWSSS Purification Method Affinity purified using the PrEST antigen as affinity ligand Verified Species Human Reactivity Recommended IHC (Immunohistochemistry) Applications - Antibody dilution: 1:200 - 1:500 - Retrieval method: HIER pH6 ICC-IF (Immunofluorescence) - Fixation/Permeabilization: PFA/Triton X-100 - Working concentration: 0.25-2 µg/ml Characterization Data Available at atlasantibodies.com/products/HPA007039 Buffer 40% glycerol and PBS (pH 7.2). 0.02% sodium azide is added as preservative. Concentration Lot dependent Storage Store at +4°C for short term storage. Long time storage is recommended at -20°C. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. For protocols, additional product information, such as images and references, see atlasantibodies.com. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -

Supplemental Material 1
Cdk20 Csrp2bp Bub1 Tmed3 1700012B15Rik Uck1 Atad2 Pbk Rfc4 Ucp3 Mtmr7 Bid Rad9 Hadh Zc3h12a Bcl2l11 1700052K11Rik Cd83 Ncaph Cdca7l Ezh2 Kif23 Rel Cdh13 Nr4a1 Pola1 Phf19 Vldlr Snx7 Fam96a Ripk2 Ccne2 Ncapd2 Pqlc3 A130009I22Rik Niacr1 Peli1 Cenpn Ralgds Dbf4 Trpc6 Gadd45b Dyrk2 Il1b Prim1 Pcna Dnmt1 Ttc30a1 Cflar Fas Tro Tank Rapgef2 Icosl 9930012K11Rik Cd28 Ccdc86 Ube2f Klhl6 Commd8 Ccrl2 D4Wsu53e Tnfaip8l2 Ehd1 Kdm2b Uhrf1 Trim37 Tecrl AI467606 Ilf2 Cobra1 Aurkb Cdc42ep2 Nfkbil2 Appbp2 Nfkbiz Zfp85-rs1 Nlrp3 Zmym5 Il6 Foxo3 Sept3 Ets2 Serpinb2 Smc3 Msh6 Itga5 Gpsm3 Slbp Dennd4c Rffl Trappc1 Mrpl34 Socs3 Ocel1 Tnfaip3 Swap70 1110003O08Rik Junb Pmm1 Cxcl10 Brca1 Acpl2 Coq10b Marcksl1 Map3k10 Braf Tnf Icam1 Pold3 AI462493 Nfkbia Dnmbp Dnajc9 Tnip1 Chfr Trim13 Slc35c2 Rb1 C78513 Limk1 C1qa Fen1 BC088983 Tsc22d3 Tnfsf9 Mapk3 E2f1 Zc3h12c Casp4 Ttf2 Maff Gmnn Zfp715 Mdm2 Mcm7 Irf1 Parp1 Ftsj2 Myo10 Irak2 Mid1ip1 Apeh Rad51c Map3k4 Tk1 E2f7 Tiparp Ptgs2 Spata13 Hmga2-ps1 Wdhd1 Clspn Ubac1 Erp29 Thap4 Dna2 Prkag2 Il1a Rfwd3 Gbp3 Cdca5 3110043O21Rik Clec4e Cxcl11 Speg Tarbp2 Cdk2 Zufsp Cdca2 E2f2 1700054N08Rik Tnfsf10 5730499H23Rik Inf2 C1qb Anapc5 Sco1 Bst2 2010107G23Rik Cnpy4 D10Wsu102e Brip1 H2-D1 Gpd1l Suv420h2 4930579G24Rik Cdc6 Cntnap2 Cyp2b10 Plod2 Irf7 C2 Lsm3 9330175E14Rik Aak1 Ick Rassf7 Cd22 Cd72 Tap1 Tcn2 Malt1 Uba7 Atp1a2 Ar Tap2 Cd86 Herc5 Heca Atp2a2 Ttc28 Dhrs3 Stat1 Ccdc22 Abca5 Nhlrc2 Ddx58 Gpx1 Dgkd Fbxl7 Lap3 Lama4 Neil1 Enpp4 Tpst1 Cybb Ccne1 Sec24d Fcgr4 Hk3 Rasgrp2 Dcun1d2 Vegfa Ecsit Ddx60 Eif2ak2 Cyp2r1 Gimap4 Fip1l1 -

Differential and Overlapping Immune Programs Regulated by IRF3 and IRF5 in Plasmacytoid Dendritic Cells
Differential and Overlapping Immune Programs Regulated by IRF3 and IRF5 in Plasmacytoid Dendritic Cells This information is current as Kwan T. Chow, Courtney Wilkins, Miwako Narita, Richard of September 28, 2021. Green, Megan Knoll, Yueh-Ming Loo and Michael Gale, Jr. J Immunol published online 8 October 2018 http://www.jimmunol.org/content/early/2018/10/05/jimmun ol.1800221 Downloaded from Supplementary http://www.jimmunol.org/content/suppl/2018/10/05/jimmunol.180022 Material 1.DCSupplemental http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication by guest on September 28, 2021 *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2018 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Published October 8, 2018, doi:10.4049/jimmunol.1800221 The Journal of Immunology Differential and Overlapping Immune Programs Regulated by IRF3 and IRF5 in Plasmacytoid Dendritic Cells Kwan T. Chow,*,† Courtney Wilkins,* Miwako Narita,‡ Richard Green,* Megan Knoll,* Yueh-Ming Loo,* and Michael Gale, Jr.* We examined the signaling pathways and cell type–specific responses of IFN regulatory factor (IRF) 5, an immune-regulatory transcription factor. -
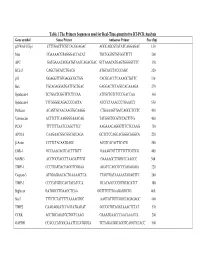
Table 1 the Primers Sequences Used for Real-Time Quantitative RT-PCR
Table 1 The Primers Sequences used for Real-Time quantitative RT-PCR Analysis Gene symbol Sense Primer Antisense Primer Size (bp) p21WAF1/Cip1 CTTTGACTTCGTCACGGAGAC AGGCAGCGTATATCAGGAGAC 150 Max CGAAAACGTAGGGACCACAT TGCTGGTGTGTGGTTTTT 200 APC GATGAAATAGGATGTAATCAGACGAC GCTAAACATGAGTGGGGTCTC 350 BCL-2 CAGCTGCACCTGACG ATGCACCTACCCAGC 320 p53 GGAGGTTGTGAGGCGCTGG CACGCACCTCAAAGCTGTTC 330 Bax TGCAGAGGATGATTGCTGAC GAGGACTCCAGCCACAAAGA 270 Syndecan-1 TCTGACTCGGTTTCTCCAA ATTGCTGTCTCCCGACCAA 360 Syndecan-3 TTTGGGGCAGACCCCACTA ATCCCTAAACCCTGAACCT 550 Perlecan ACAGTGCAACAAGTGCAAGG CTGAAAGTGACCAGGCTCCTC 450 Vitronectin ACTTCTTCAAGGGGAAACAG TATGGCTGCGTCCACTTTG 400 PCAF TTTTTTTAATCCAGCTTCC AAGAAACAGGGTTTCTCCAAG 750 APO1A CAAGAACGGCGGCGCCAGA GCTCTCCAGCACGGGCAGGCA 220 β-Actin CCTTCTACAATGAGC ACGTCACACTTCATG 260 EGR-1 GCCAAACAGTCACTTTGTT GAAAGTGTTTTTTCTTCGTCG 400 MAPK3 ACCTGCGACCTTAAGATTTGT GAAAAGCTTGGCCCAAGCC 300 TIMP-1 CCCTGATGACGAGGTCGGAA AGATCCAGCGCCCAGAGAGA 220 Caspase-3 ATGGAGAACACTGAAAACTCA TTAGTGATAAAAATAGAGTTC 200 TIMP-3 CCCCATGTGCAGTACATCCA GCACAGCCCCGTGTACATCT 180 Biglycan GATGGCCTGAAGCTCAA GGTTTGTTGAAGAGGCTG 460 Stat3 TTTCTCTATTTTTAAAAGTGC AAGTATTGTCGGCCAGAGAGC 460 TIMP2 CAAGAGGATCCAGTATGAGAT GCCCGTGTAGATAAACTCTAT 570 CCRK ACCTGCAGATGCTGCTCAAG GAAATGAACCCAACAAACCA 200 GAPDH CCACCCATGGCAAATTCCATGGGA TCTAGACGGCAGGTCAGGTCCACC 500 Table 2. Genes with Altered Expression in Cleaved vs. Native Antithrombin-Treated HUVECs Gene Fold Changes Gene Symbol Gene Description Cluster ID (ATC/ATN) Adhesion/ECM ARGN Agrin Hs.273330 2.18 CDH24 Caderhrin-like -

Molecular Signatures of Membrane Protein Complexes Underlying Muscular Dystrophy*□S
crossmark Research Author’s Choice © 2016 by The American Society for Biochemistry and Molecular Biology, Inc. This paper is available on line at http://www.mcponline.org Molecular Signatures of Membrane Protein Complexes Underlying Muscular Dystrophy*□S Rolf Turk‡§¶ʈ**, Jordy J. Hsiao¶, Melinda M. Smits¶, Brandon H. Ng¶, Tyler C. Pospisil‡§¶ʈ**, Kayla S. Jones‡§¶ʈ**, Kevin P. Campbell‡§¶ʈ**, and Michael E. Wright¶‡‡ Mutations in genes encoding components of the sar- The muscular dystrophies are hereditary diseases charac- colemmal dystrophin-glycoprotein complex (DGC) are re- terized primarily by the progressive degeneration and weak- sponsible for a large number of muscular dystrophies. As ness of skeletal muscle. Most are caused by deficiencies in such, molecular dissection of the DGC is expected to both proteins associated with the cell membrane (i.e. the sarco- reveal pathological mechanisms, and provides a biologi- lemma in skeletal muscle), and typical features include insta- cal framework for validating new DGC components. Es- bility of the sarcolemma and consequent death of the myofi- tablishment of the molecular composition of plasma- ber (1). membrane protein complexes has been hampered by a One class of muscular dystrophies is caused by mutations lack of suitable biochemical approaches. Here we present in genes that encode components of the sarcolemmal dys- an analytical workflow based upon the principles of pro- tein correlation profiling that has enabled us to model the trophin-glycoprotein complex (DGC). In differentiated skeletal molecular composition of the DGC in mouse skeletal mus- muscle, this structure links the extracellular matrix to the cle. We also report our analysis of protein complexes in intracellular cytoskeleton. -

PRODUCTS and SERVICES Target List
PRODUCTS AND SERVICES Target list Kinase Products P.1-11 Kinase Products Biochemical Assays P.12 "QuickScout Screening Assist™ Kits" Kinase Protein Assay Kits P.13 "QuickScout Custom Profiling & Panel Profiling Series" Targets P.14 "QuickScout Custom Profiling Series" Preincubation Targets Cell-Based Assays P.15 NanoBRET™ TE Intracellular Kinase Cell-Based Assay Service Targets P.16 Tyrosine Kinase Ba/F3 Cell-Based Assay Service Targets P.17 Kinase HEK293 Cell-Based Assay Service ~ClariCELL™ ~ Targets P.18 Detection of Protein-Protein Interactions ~ProbeX™~ Stable Cell Lines Crystallization Services P.19 FastLane™ Structures ~Premium~ P.20-21 FastLane™ Structures ~Standard~ Kinase Products For details of products, please see "PRODUCTS AND SERVICES" on page 1~3. Tyrosine Kinases Note: Please contact us for availability or further information. Information may be changed without notice. Expression Protein Kinase Tag Carna Product Name Catalog No. Construct Sequence Accession Number Tag Location System HIS ABL(ABL1) 08-001 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) BTN BTN-ABL(ABL1) 08-401-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ABL(ABL1) [E255K] HIS ABL(ABL1)[E255K] 08-094 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) HIS ABL(ABL1)[T315I] 08-093 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) [T315I] BTN BTN-ABL(ABL1)[T315I] 08-493-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ACK(TNK2) GST ACK(TNK2) 08-196 Catalytic domain -

Supplementary Table 5.List of the 220 Most Frequently Amplified Genes In
Supplementary Table 5. List of the 220 most frequently amplified genes in this study. The table includes their chromosomal location, the amplification frequency in ER-positive female breast cancer with associated p-value for difference in proportions, the preference for surrogate intrinsic molecular subtype, and associations with clinical, pathological and genetic characteristics. Potentially druggable gene categories, clinical actionability and known drug interactions are indicated per gene. Gene Full name chr location % amp in FFPE % amps in FF total % amp % amp ER+ FBC* p-value MBC vs ER+ FBC** % in lumA-like % in lumB-like p-value BRCA2 germline Age Hist type ER status PR status HER2 status Grade MAI Size LN SNV load PIK3CA mut KM (OS)*** KM (5Y OS)*** druggable gene category# clinically actionable?## known drug interactions?### THBS1 thrombospondin 1 15q14 37% 9% 30% 0.1% <0.0001 23% 35% 0.128 ns ns ns ns ns ns ns ns ns ns ns ns 0.642 p=0.832 cell surface, tumor suppressor, drug resistance, external side of plasma membrane no none PRKDC protein kinase, DNA-activated, catalytic polypeptide 8q11.21 35% 7% 27% 10.9% <0.0001 26% 30% 0.595 ns ns ns ns ns ns ns ns ns ns ns ns 0.838 p=0.903 (serine threonine) kinase, druggable genome, PI3 kinase, tumor suppressor, TF complex, TF binding, DNA repair yes DNA-PK INHIBITOR V (DNA-PK inhibitor); WORTMANNIN (PI3K inhibitor); SF1126 (PI3 kinase/mTOR inhibitor) TBX3 T-box 3 12q24.21 34% 7% 27% 0.1% <0.0001 20% 35% 0.053 ns ns ns ns ns ns ns ns ns ns ns ns 0.439 p=0.264 tumor suppressor, TF binding -

Clinical, Molecular, and Immune Analysis of Dabrafenib-Trametinib
Supplementary Online Content Chen G, McQuade JL, Panka DJ, et al. Clinical, molecular and immune analysis of dabrafenib-trametinib combination treatment for metastatic melanoma that progressed during BRAF inhibitor monotherapy: a phase 2 clinical trial. JAMA Oncology. Published online April 28, 2016. doi:10.1001/jamaoncol.2016.0509. eMethods. eReferences. eTable 1. Clinical efficacy eTable 2. Adverse events eTable 3. Correlation of baseline patient characteristics with treatment outcomes eTable 4. Patient responses and baseline IHC results eFigure 1. Kaplan-Meier analysis of overall survival eFigure 2. Correlation between IHC and RNAseq results eFigure 3. pPRAS40 expression and PFS eFigure 4. Baseline and treatment-induced changes in immune infiltrates eFigure 5. PD-L1 expression eTable 5. Nonsynonymous mutations detected by WES in baseline tumors This supplementary material has been provided by the authors to give readers additional information about their work. © 2016 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 09/30/2021 eMethods Whole exome sequencing Whole exome capture libraries for both tumor and normal samples were constructed using 100ng genomic DNA input and following the protocol as described by Fisher et al.,3 with the following adapter modification: Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter. In-solution hybrid selection was performed using the Illumina Rapid Capture Exome enrichment kit with 38Mb target territory (29Mb baited). The targeted region includes 98.3% of the intervals in the Refseq exome database. Dual-indexed libraries were pooled into groups of up to 96 samples prior to hybridization.