Supplementary Table 5.List of the 220 Most Frequently Amplified Genes In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Download (Pdf)
Invivoscribe's wholly-owned Laboratories for Personalized Molecular LabPMM LLC Medicine® (LabPMM) is a network of international reference laboratories that provide the medical and pharmaceutical communities with worldwide Located in San Diego, California, USA, it holds access to harmonized and standardized clinical testing services. We view the following accreditations and certifications: internationally reproducible and concordant testing as a requirement for ISO 15189, CAP, and CLIA, and is licensed to provide diagnostic consistent stratification of patients for enrollment in clinical trials, and the laboratory services in the states of California, Florida, foundation for establishing optimized treatment schedules linked to patient’s Maryland, New York, Pennsylvania, and Rhode Island. individual profile. LabPMM provides reliable patient stratification at diagnosis LabPMM GmbH and monitoring, throughout the entire course of treatment in support of Personalized Molecular Medicine® and Personalized Based in Martinsried (Munich), Germany. It is an ISO 15189 Molecular Diagnostics®. accredited international reference laboratory. CLIA/CAP accreditation is planned. Invivoscribe currently operates four clinical laboratories to serve partners in the USA (San Diego, CA), Europe (Munich, Germany), and Asia (Tokyo, Japan and Shanghai, China). These laboratories use the same critical LabPMM 合同会社 reagents and software which are developed consistently with ISO Located in Kawasaki (Tokyo), Japan and a licensed clinical lab. 13485 design control. Our cGMP reagents, rigorous standards for assay development & validation, and testing performed consistently under ISO 15189 requirements help ensure LabPMM generates standardized and concordant test results worldwide. Invivoscribe Diagnostic Technologies (Shanghai) Co., Ltd. LabPMM is an international network of PersonalMed Laboratories® focused on molecular oncology biomarker studies. Located in Shangai, China. -

Systematic Screening for Potential Therapeutic Targets in Osteosarcoma Through a Kinome-Wide CRISPR-Cas9 Library
Cancer Biol Med 2020. doi: 10.20892/j.issn.2095-3941.2020.0162 ORIGINAL ARTICLE Systematic screening for potential therapeutic targets in osteosarcoma through a kinome-wide CRISPR-Cas9 library Yuanzhong Wu*, Liwen Zhou*, Zifeng Wang, Xin Wang, Ruhua Zhang, Lisi Zheng, Tiebang Kang Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, China ABSTRACT Objective: Osteosarcoma is the most common primary malignant bone tumor. However, the survival of patients with osteosarcoma has remained unchanged during the past 30 years, owing to a lack of efficient therapeutic targets. Methods: We constructed a kinome-targeting CRISPR-Cas9 library containing 507 kinases and 100 nontargeting controls and screened the potential kinase targets in osteosarcoma. The CRISPR screening sequencing data were analyzed with the Model-based Analysis of Genome-wide CRISPR/Cas9 Knockout (MAGeCK) Python package. The functional data were applied in the 143B cell line through lenti-CRISPR-mediated gene knockout. The clinical significance of kinases in the survival of patients with osteosarcoma was analyzed in the R2: Genomics Analysis and Visualization Platform. Results: We identified 53 potential kinase targets in osteosarcoma. Among these targets, we analyzed 3 kinases, TRRAP, PKMYT1, and TP53RK, to validate their oncogenic functions in osteosarcoma. PKMYT1 and TP53RK showed higher expression in osteosarcoma than in normal bone tissue, whereas TRRAP showed no significant difference. High expression of all 3 kinases was associated with relatively poor prognosis in patients with osteosarcoma. Conclusions: Our results not only offer potential therapeutic kinase targets in osteosarcoma but also provide a paradigm for functional genetic screening by using a CRISPR-Cas9 library, including target design, library construction, screening workflow, data analysis, and functional validation. -

Calpain-5 Gene Expression in the Mouse Eye and Brain
Schaefer et al. BMC Res Notes (2017) 10:602 DOI 10.1186/s13104-017-2927-8 BMC Research Notes RESEARCH NOTE Open Access Calpain‑5 gene expression in the mouse eye and brain Kellie Schaefer1, MaryAnn Mahajan1, Anuradha Gore1, Stephen H. Tsang3, Alexander G. Bassuk4 and Vinit B. Mahajan1,2* Abstract Objective: Our objective was to characterize CAPN5 gene expression in the mouse central nervous system. Mouse brain and eye sections were probed with two high-afnity RNA oligonucleotide analogs designed to bind CAPN5 RNA and one scramble, control oligonucleotide. Images were captured in brightfeld. Results: CAPN5 RNA probes were validated on mouse breast cancer tumor tissue. In the eye, CAPN5 was expressed in the ganglion cell, inner nuclear and outer nuclear layers of the retina. Signal could not be detected in the ciliary body or the iris because of the high density of melanin. In the brain, CAPN5 was expressed in the granule cell layers of the hippocampus and cerebellum. There was scattered expression in pons. The visual cortex showed faint signal. Most signal in the brain was in a punctate pattern. Keywords: CAPN5, Calpain, In situ hybridization, Retina, Brain, Gene expression Introduction pigmentosa, retinal neovascularization, and proliferative Calpain-5 (CAPN5) is a member of the calpain family of retinopathy. Which ultimately leads to blindness [20]. calcium-activated proteases that target a variety of path- Currently there is no treatment. ways to exert control over numerous processes, includ- An important question to understanding how CAPN5 ing tissue necrosis, cytoskeletal remodeling, cell-cycle leads to disease is identifying which tissues CAPN5 is control, cell migration, myofbril turnover, regulation expressed in and the levels of CAPN5 in those tissues. -

The Role of the Mtor Pathway in Developmental Reprogramming Of
THE ROLE OF THE MTOR PATHWAY IN DEVELOPMENTAL REPROGRAMMING OF HEPATIC LIPID METABOLISM AND THE HEPATIC TRANSCRIPTOME AFTER EXPOSURE TO 2,2',4,4'- TETRABROMODIPHENYL ETHER (BDE-47) An Honors Thesis Presented By JOSEPH PAUL MCGAUNN Approved as to style and content by: ________________________________________________________** Alexander Suvorov 05/18/20 10:40 ** Chair ________________________________________________________** Laura V Danai 05/18/20 10:51 ** Committee Member ________________________________________________________** Scott C Garman 05/18/20 10:57 ** Honors Program Director ABSTRACT An emerging hypothesis links the epidemic of metabolic diseases, such as non-alcoholic fatty liver disease (NAFLD) and diabetes with chemical exposures during development. Evidence from our lab and others suggests that developmental exposure to environmentally prevalent flame-retardant BDE47 may permanently reprogram hepatic lipid metabolism, resulting in an NAFLD-like phenotype. Additionally, we have demonstrated that BDE-47 alters the activity of both mTOR complexes (mTORC1 and 2) in hepatocytes. The mTOR pathway integrates environmental information from different signaling pathways, and regulates key cellular functions such as lipid metabolism, innate immunity, and ribosome biogenesis. Thus, we hypothesized that the developmental effects of BDE-47 on liver lipid metabolism are mTOR-dependent. To assess this, we generated mice with liver-specific deletions of mTORC1 or mTORC2 and exposed these mice and their respective controls perinatally to -

AGC Kinases in Mtor Signaling, in Mike Hall and Fuyuhiko Tamanoi: the Enzymes, Vol
Provided for non-commercial research and educational use only. Not for reproduction, distribution or commercial use. This chapter was originally published in the book, The Enzymes, Vol .27, published by Elsevier, and the attached copy is provided by Elsevier for the author's benefit and for the benefit of the author's institution, for non-commercial research and educational use including without limitation use in instruction at your institution, sending it to specific colleagues who know you, and providing a copy to your institution’s administrator. All other uses, reproduction and distribution, including without limitation commercial reprints, selling or licensing copies or access, or posting on open internet sites, your personal or institution’s website or repository, are prohibited. For exceptions, permission may be sought for such use through Elsevier's permissions site at: http://www.elsevier.com/locate/permissionusematerial From: ESTELA JACINTO, AGC Kinases in mTOR Signaling, In Mike Hall and Fuyuhiko Tamanoi: The Enzymes, Vol. 27, Burlington: Academic Press, 2010, pp.101-128. ISBN: 978-0-12-381539-2, © Copyright 2010 Elsevier Inc, Academic Press. Author's personal copy 7 AGC Kinases in mTOR Signaling ESTELA JACINTO Department of Physiology and Biophysics UMDNJ-Robert Wood Johnson Medical School, Piscataway New Jersey, USA I. Abstract The mammalian target of rapamycin (mTOR), a protein kinase with homology to lipid kinases, orchestrates cellular responses to growth and stress signals. Various extracellular and intracellular inputs to mTOR are known. mTOR processes these inputs as part of two mTOR protein com- plexes, mTORC1 or mTORC2. Surprisingly, despite the many cellular functions that are linked to mTOR, there are very few direct mTOR substrates identified to date. -

Advancing a Clinically Relevant Perspective of the Clonal Nature of Cancer
Advancing a clinically relevant perspective of the clonal nature of cancer Christian Ruiza,b, Elizabeth Lenkiewicza, Lisa Eversa, Tara Holleya, Alex Robesona, Jeffrey Kieferc, Michael J. Demeurea,d, Michael A. Hollingsworthe, Michael Shenf, Donna Prunkardf, Peter S. Rabinovitchf, Tobias Zellwegerg, Spyro Moussesc, Jeffrey M. Trenta,h, John D. Carpteni, Lukas Bubendorfb, Daniel Von Hoffa,d, and Michael T. Barretta,1 aClinical Translational Research Division, Translational Genomics Research Institute, Scottsdale, AZ 85259; bInstitute for Pathology, University Hospital Basel, University of Basel, 4031 Basel, Switzerland; cGenetic Basis of Human Disease, Translational Genomics Research Institute, Phoenix, AZ 85004; dVirginia G. Piper Cancer Center, Scottsdale Healthcare, Scottsdale, AZ 85258; eEppley Institute for Research in Cancer and Allied Diseases, Nebraska Medical Center, Omaha, NE 68198; fDepartment of Pathology, University of Washington, Seattle, WA 98105; gDivision of Urology, St. Claraspital and University of Basel, 4058 Basel, Switzerland; hVan Andel Research Institute, Grand Rapids, MI 49503; and iIntegrated Cancer Genomics Division, Translational Genomics Research Institute, Phoenix, AZ 85004 Edited* by George F. Vande Woude, Van Andel Research Institute, Grand Rapids, MI, and approved June 10, 2011 (received for review March 11, 2011) Cancers frequently arise as a result of an acquired genomic insta- on the basis of morphology alone (8). Thus, the application of bility and the subsequent clonal evolution of neoplastic cells with purification methods such as laser capture microdissection does variable patterns of genetic aberrations. Thus, the presence and not resolve the complexities of many samples. A second approach behaviors of distinct clonal populations in each patient’s tumor is to passage tumor biopsies in tissue culture or in xenografts (4, 9– may underlie multiple clinical phenotypes in cancers. -

Profiling Data
Compound Name DiscoveRx Gene Symbol Entrez Gene Percent Compound Symbol Control Concentration (nM) JNK-IN-8 AAK1 AAK1 69 1000 JNK-IN-8 ABL1(E255K)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317I)-nonphosphorylated ABL1 87 1000 JNK-IN-8 ABL1(F317I)-phosphorylated ABL1 100 1000 JNK-IN-8 ABL1(F317L)-nonphosphorylated ABL1 65 1000 JNK-IN-8 ABL1(F317L)-phosphorylated ABL1 61 1000 JNK-IN-8 ABL1(H396P)-nonphosphorylated ABL1 42 1000 JNK-IN-8 ABL1(H396P)-phosphorylated ABL1 60 1000 JNK-IN-8 ABL1(M351T)-phosphorylated ABL1 81 1000 JNK-IN-8 ABL1(Q252H)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(Q252H)-phosphorylated ABL1 56 1000 JNK-IN-8 ABL1(T315I)-nonphosphorylated ABL1 100 1000 JNK-IN-8 ABL1(T315I)-phosphorylated ABL1 92 1000 JNK-IN-8 ABL1(Y253F)-phosphorylated ABL1 71 1000 JNK-IN-8 ABL1-nonphosphorylated ABL1 97 1000 JNK-IN-8 ABL1-phosphorylated ABL1 100 1000 JNK-IN-8 ABL2 ABL2 97 1000 JNK-IN-8 ACVR1 ACVR1 100 1000 JNK-IN-8 ACVR1B ACVR1B 88 1000 JNK-IN-8 ACVR2A ACVR2A 100 1000 JNK-IN-8 ACVR2B ACVR2B 100 1000 JNK-IN-8 ACVRL1 ACVRL1 96 1000 JNK-IN-8 ADCK3 CABC1 100 1000 JNK-IN-8 ADCK4 ADCK4 93 1000 JNK-IN-8 AKT1 AKT1 100 1000 JNK-IN-8 AKT2 AKT2 100 1000 JNK-IN-8 AKT3 AKT3 100 1000 JNK-IN-8 ALK ALK 85 1000 JNK-IN-8 AMPK-alpha1 PRKAA1 100 1000 JNK-IN-8 AMPK-alpha2 PRKAA2 84 1000 JNK-IN-8 ANKK1 ANKK1 75 1000 JNK-IN-8 ARK5 NUAK1 100 1000 JNK-IN-8 ASK1 MAP3K5 100 1000 JNK-IN-8 ASK2 MAP3K6 93 1000 JNK-IN-8 AURKA AURKA 100 1000 JNK-IN-8 AURKA AURKA 84 1000 JNK-IN-8 AURKB AURKB 83 1000 JNK-IN-8 AURKB AURKB 96 1000 JNK-IN-8 AURKC AURKC 95 1000 JNK-IN-8 -

The Role of Cyclin B3 in Mammalian Meiosis
THE ROLE OF CYCLIN B3 IN MAMMALIAN MEIOSIS by Mehmet Erman Karasu A Dissertation Presented to the Faculty of the Louis V. Gerstner Jr. Graduate School of Biomedical Sciences, Memorial Sloan Kettering Cancer Center In Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy New York, NY November, 2018 Scott Keeney, PhD Date Dissertation Mentor Copyright © Mehmet Erman Karasu 2018 DEDICATION I would like to dedicate this thesis to my parents, Mukaddes and Mustafa Karasu. I have been so lucky to have their support and unconditional love in this life. ii ABSTRACT Cyclins and cyclin dependent kinases (CDKs) lie at the center of the regulation of the cell cycle. Cyclins as regulatory partners of CDKs control the switch-like cell cycle transitions that orchestrate orderly duplication and segregation of genomes. Similar to somatic cell division, temporal regulation of cyclin-CDK activity is also important in meiosis, which is the specialized cell division that generates gametes for sexual production by halving the genome. Meiosis does so by carrying out one round of DNA replication followed by two successive divisions without another intervening phase of DNA replication. In budding yeast, cyclin-CDK activity has been shown to have a crucial role in meiotic events such as formation of meiotic double-strand breaks that initiate homologous recombination. Mammalian cells express numerous cyclins and CDKs, but how these proteins control meiosis remains poorly understood. Cyclin B3 was previously identified as germ cell specific, and its restricted expression pattern at the beginning of meiosis made it an interesting candidate to regulate meiotic events. -
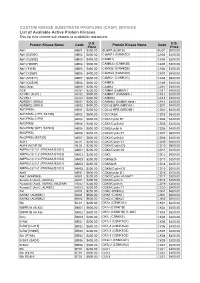
CUSTOM KINASE SUBSTRATE PROFILING (CKSP) SERVICE List of Available Active Protein Kinases This List May Change with Respect to Availability and Pricing
CUSTOM KINASE SUBSTRATE PROFILING (CKSP) SERVICE List of Available Active Protein Kinases This list may change with respect to availability and pricing. Protein Kinase Name Code U.S. Protein Kinase Name Code U.S. Price Price Abl1 AB01 $200.00 BUBR1(BUB1B) BU01 $600.00 Abl1 [E255K] AB02 $400.00 CaMK1δ (CAMK1D) CA03 $200.00 Abl1 [G250E] AB03 $400.00 CAMK1γ CA04 $200.00 Abl1 [H369P] AB04 $600.00 CAMK2α (CAMK2B) CA05 $200.00 Abl1 [T315I] AB05 $400.00 CaMK2β (CAMK2B) CA06 $200.00 Abl1 [Y253F] AB06 $400.00 CaMK2δ (CAMK2D) CA07 $400.00 Abl1 [M351T] AB07 $600.00 CaMK2γ (CAMK2G) CA08 $600.00 Abl1 [Q252H] AB08 $600.00 CAMK3γ CA09 $200.00 Abl2 (Arg) AB09 $200.00 CAMK4 CA10 $200.00 ACK AC01 $200.00 CAMK4 (CaMKIV ) CA11 $400.00 ACVR1 (ALK2 ) AC02 $400.00 CAMKK1 (CAMKKA ) CA12 $200.00 ACVRL1 AC03 $400.00 CAMKK2 CA13 $200.00 ADRBK1 (GRK2) AD01 $200.00 CAMKK2 (CaMKK beta ) CA14 $400.00 ADRBK2 (GRK3) AD02 $400.00 CDC42 BPA (MRCKA ) CD01 $400.00 Akt1/PKBα AK01 $200.00 CDC42 BPB (MRCKB ) CD02 $400.00 Akt1/PKBα [δPH, S473D] AK02 $600.00 CDC7/ASK CD03 $600.00 Akt1/PKBα [δPH] AK03 $600.00 CDK1/cyclin B1 CD04 $400.00 Akt2/PKBβ AK04 $200.00 CDK1/CyclinA2 CD05 $200.00 Akt2/PKBβ [δPH, S474D] AK05 $600.00 CDK2/cyclin A CD06 $400.00 Akt3/PKBγ AK06 $200.00 CDK2/Cyclin E1 CD07 $600.00 Akt3/PKBγ [S472D] AK07 $600.00 CDK2/CyclinA2 CD08 $200.00 ALK1 AL01 $200.00 CDK3/Cyclin E1 CD09 $600.00 ALK4 (ACVR1B) AL02 $200.00 CDK4//Cyclin D3 CD10 $600.00 AMPKα1/β1/γ1 (PRKAA1/B1/G1) AM01 $200.00 CDK4/Cyclin D1 CD11 $200.00 AMPKα1/β1/γ2 (PRKAA1/B1/G2) AM02 $200.00 CDK5 CD12 $600.00 -

Src-Family Kinases Impact Prognosis and Targeted Therapy in Flt3-ITD+ Acute Myeloid Leukemia
Src-Family Kinases Impact Prognosis and Targeted Therapy in Flt3-ITD+ Acute Myeloid Leukemia Title Page by Ravi K. Patel Bachelor of Science, University of Minnesota, 2013 Submitted to the Graduate Faculty of School of Medicine in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2019 Commi ttee Membership Pa UNIVERSITY OF PITTSBURGH SCHOOL OF MEDICINE Commi ttee Membership Page This dissertation was presented by Ravi K. Patel It was defended on May 31, 2019 and approved by Qiming (Jane) Wang, Associate Professor Pharmacology and Chemical Biology Vaughn S. Cooper, Professor of Microbiology and Molecular Genetics Adrian Lee, Professor of Pharmacology and Chemical Biology Laura Stabile, Research Associate Professor of Pharmacology and Chemical Biology Thomas E. Smithgall, Dissertation Director, Professor and Chair of Microbiology and Molecular Genetics ii Copyright © by Ravi K. Patel 2019 iii Abstract Src-Family Kinases Play an Important Role in Flt3-ITD Acute Myeloid Leukemia Prognosis and Drug Efficacy Ravi K. Patel, PhD University of Pittsburgh, 2019 Abstract Acute myelogenous leukemia (AML) is a disease characterized by undifferentiated bone-marrow progenitor cells dominating the bone marrow. Currently the five-year survival rate for AML patients is 27.4 percent. Meanwhile the standard of care for most AML patients has not changed for nearly 50 years. We now know that AML is a genetically heterogeneous disease and therefore it is unlikely that all AML patients will respond to therapy the same way. Upregulation of protein-tyrosine kinase signaling pathways is one common feature of some AML tumors, offering opportunities for targeted therapy. -

Autophagy Processes Are Dependent on EGF Receptor Signaling
www.oncotarget.com Oncotarget, 2018, Vol. 9, (No. 54), pp: 30289-30303 Research Paper Autophagy processes are dependent on EGF receptor signaling Vincenzo De Iuliis1, Antonio Marino1, Marika Caruso1, Sabrina Capodifoglio1, Vincenzo Flati2, Anna Marynuk3, Valeria Marricareda3, Sebastiano Ursi1, Paola Lanuti4, Claudio Talora5, Pio Conti6, Stefano Martinotti1,7 and Elena Toniato7 1Unit of Predictive Medicine, SS Annunziata University Hospital of Chieti, Chieti, Italy 2Department of Biotechnological and Applied Clinical Sciences, University of L’Aquila, L’Aquila, Italy 3Odessa National Medical University, Odesa, Odessa Oblsat, Ucraina 4Department of Medicine and Aging Sciences, University G. d’Annunzio of Chieti, Chieti, Italy 5Department of Molecular Medicine, University of Rome “La Sapienza”, Rome, Italy 6Postgraduate Medical School, University of Chieti, Chieti, Italy 7Department of Medical, Oral and Biotechnological Sciences, University G. d’Annunzio of Chieti, Chieti, Italy Correspondence to: Elena Toniato, email: [email protected] Keywords: apoptosis; autophagy; Beclin 1; EGF receptor; MAPK pathway Received: May 03, 2018 Accepted: June 13, 2018 Published: July 13, 2018 Copyright: De Iuliis et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Autophagy is a not well-understood conserved mechanism activated during nutritional deprivation in order to maintain cellular homeostasis. In the present study, we investigated the correlations between autophagy, apoptosis and the MAPK pathways in melanoma cell lines. We demonstrated that during starvation the EGF receptor mediated signaling activates many proteins involved in the MAPK pathway.