Rozina Aslam Biochemistry UA
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Citrus from Seed?
Which citrus fruits will come true to type Orogrande, Tomatera, Fina, Nour, Hernandina, Clementard.) from seed? Ellendale Tom McClendon writes in Hardy Citrus Encore for the South East: Fortune Fremont (50% monoembryonic) “Most common citrus such as oranges, Temple grapefruit, lemons and most mandarins Ugli Umatilla are polyembryonic and will come true to Wilking type. Because most citrus have this trait, Highly polyembryonic citrus types : will mostly hybridization can be very difficult to produce nucellar polyembryonic seeds that will grow true to type. achieve…. This unique characteristic Citrus × aurantiifolia Mexican lime (Key lime, West allows amateurs to grow citrus from seed, Indian lime) something you can’t do with, say, Citrus × insitorum (×Citroncirus webberii) Citranges, such as Rusk, Troyer etc. apples.” [12*] Citrus × jambhiri ‘Rough lemon’, ‘Rangpur’ lime, ‘Otaheite’ lime Monoembryonic (don’t come true) Citrus × limettioides Palestine lime (Indian sweet lime) Citrus × microcarpa ‘Calamondin’ Meyer Lemon Citrus × paradisi Grapefruit (Marsh, Star Ruby, Nagami Kumquat Redblush, Chironja, Smooth Flat Seville) Marumi Kumquat Citrus × sinensis Sweet oranges (Blonde, navel and Pummelos blood oranges) Temple Tangor Citrus amblycarpa 'Nasnaran' mandarin Clementine Mandarin Citrus depressa ‘Shekwasha’ mandarin Citrus karna ‘Karna’, ‘Khatta’ Poncirus Trifoliata Citrus kinokuni ‘Kishu mandarin’ Citrus lycopersicaeformis ‘Kokni’ or ‘Monkey mandarin’ Polyembryonic (come true) Citrus macrophylla ‘Alemow’ Most Oranges Citrus reshni ‘Cleopatra’ mandarin Changshou Kumquat Citrus sunki (Citrus reticulata var. austera) Sour mandarin Meiwa Kumquat (mostly polyembryonic) Citrus trifoliata (Poncirus trifoliata) Trifoliate orange Most Satsumas and Tangerines The following mandarin varieties are polyembryonic: Most Lemons Dancy Most Limes Emperor Grapefruits Empress Tangelos Fairchild Kinnow Highly monoembryonic citrus types: Mediterranean (Avana, Tardivo di Ciaculli) Will produce zygotic monoembryonic seeds that will not Naartje come true to type. -
2019 Full Provisional List
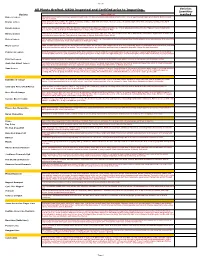
Sheet1 All Plants Grafted. USDA inspected and Certified prior to Importing. Varieties Quantities Variety Description required Baboon Lemon A Brazilian lemon with very intense yellow rind and flesh. The flavour is acidic with almost a hint of lime. Tree is vigorous with large green leaves. Both tree and fruit are beautiful. Bearss Lemon 1952. Fruit closely resembles the Lisbon. Very juicy and has a high rind oil content. The leaves are a beautiful purple when first emerging, turning a nice dark green. Fruit is ready from June to December. Eureka Lemon Fruit is very juicy and highly acidic. The Eureka originated in Los Angeles, California and is one of their principal varieties. It is the "typical" lemon found in the grocery stores, nice yellow colour with typical lemon shape. Harvested November to May Harvey Lemon 1948.Having survived the disastrous deep freezes in Florida during the ’60’s and ’70’s. this varieties is known to withstand cold weather. Typical lemon shape and tart, juicy true lemon flavour. Fruit ripens in September to March. Self fertile. Zones 8A-10. Lisbon Lemon Fruit is very juicy and acic. The leaves are dense and tree is very vigorous. This Lisbon is more cold tolerant than the Eureka and is more productive. It is one of the major varieties in California. Fruit is harvested from February to May. Meyer Lemon 1908. Considered ever-bearing, the blooms are very aromatic. It is a lemon and orange hybrid. It is very cold hardy. Fruit is round with a thin rind. Fruit is juicy and has a very nice flavour, with a low acidity. -

Citrus Industry Biosecurity Plan 2015
Industry Biosecurity Plan for the Citrus Industry Version 3.0 July 2015 PLANT HEALTH AUSTRALIA | Citrus Industry Biosecurity Plan 2015 Location: Level 1 1 Phipps Close DEAKIN ACT 2600 Phone: +61 2 6215 7700 Fax: +61 2 6260 4321 E-mail: [email protected] Visit our web site: www.planthealthaustralia.com.au An electronic copy of this plan is available through the email address listed above. © Plant Health Australia Limited 2004 Copyright in this publication is owned by Plant Health Australia Limited, except when content has been provided by other contributors, in which case copyright may be owned by another person. With the exception of any material protected by a trade mark, this publication is licensed under a Creative Commons Attribution-No Derivs 3.0 Australia licence. Any use of this publication, other than as authorised under this licence or copyright law, is prohibited. http://creativecommons.org/licenses/by-nd/3.0/ - This details the relevant licence conditions, including the full legal code. This licence allows for redistribution, commercial and non-commercial, as long as it is passed along unchanged and in whole, with credit to Plant Health Australia (as below). In referencing this document, the preferred citation is: Plant Health Australia Ltd (2004) Industry Biosecurity Plan for the Citrus Industry (Version 3.0 – July 2015). Plant Health Australia, Canberra, ACT. Disclaimer: The material contained in this publication is produced for general information only. It is not intended as professional advice on any particular matter. No person should act or fail to act on the basis of any material contained in this publication without first obtaining specific and independent professional advice. -

Industry Biosecurity Plan
Threat specific contingency osecurity Project plan for huanglongbing and its vectors Nursery production Plant Health & Bi Nursery work: levy at Queensland Department of Agriculture, Fisheries and Forestry September 2013 Disclaimer The scientific and technical content of this document is current to the date published and all efforts have been made to obtain relevant and published information on the pest. New information will be included as it becomes available, or when the document is reviewed. The material contained in this publication is produced for general information only. It is not intended as professional advice on any particular matter. No person should act or fail to act on the basis of any material contained in this publication without first obtaining specific, independent professional advice. Plant Health Australia and all persons acting for Plant Health Australia in preparing this publication, expressly disclaim all and any liability to any persons in respect of anything done by any such person in reliance, whether in whole or in part, on this publication. The views expressed in this publication are not necessarily those of Plant Health Australia. Acknowledgements This contingency plan was prepared by Ken Pegg, Andrew Manners and Lindy Coates. Thanks go to Tony Cooke (DAFF Queensland), John McDonald (NGIQ), Andrew Miles (DAFF Queensland), Patricia Barkley (Citrus Australia Ltd.), Andrew Beattie (University of Western Sydney), Ceri Pearce and Christine Horlock (DAFF Queensland) for providing helpful comments that improved this contingency plan significantly. “Huanglongbing and its Vectors - A Pest Specific Contingency Plan for the Citrus and Nursery and Garden Industries”, which was prepared for the citrus and nursery and garden industries by Andrew Beattie and Patricia Barkley in 2009, was used extensively as a source of information in the preparation of this document. -

New and Noteworthy Citrus Varieties Presentation
New and Noteworthy Citrus Varieties Citrus species & Citrus Relatives Hundreds of varieties available. CITRON Citrus medica • The citron is believed to be one of the original kinds of citrus. • Trees are small and shrubby with an open growth habit. The new growth and flowers are flushed with purple and the trees are sensitive to frost. • Ethrog or Etrog citron is a variety of citron commonly used in the Jewish Feast of Tabernacles. The flesh is pale yellow and acidic, but not very juicy. The fruits hold well on the tree. The aromatic fruit is considerably larger than a lemon. • The yellow rind is glossy, thick and bumpy. Citron rind is traditionally candied for use in holiday fruitcake. Ethrog or Etrog citron CITRON Citrus medica • Buddha’s Hand or Fingered citron is a unique citrus grown mainly as a curiosity. The six to twelve inch fruits are apically split into a varying number of segments that are reminiscent of a human hand. • The rind is yellow and highly fragrant at maturity. The interior of the fruit is solid rind with no flesh or seeds. • Fingered citron fruits usually mature in late fall to early winter and hold moderately well on the tree, but not as well as other citron varieties. Buddha’s Hand or Fingered citron NAVEL ORANGES Citrus sinensis • ‘Washington navel orange’ is also known • ‘Lane Late Navel’ was the first of a as the Bahia. It was imported into the number of late maturing Australian United States in 1870. navel orange bud sport selections of Washington navel imported into • These exceptionally delicious, seedless, California. -

Citrus Cavaleriei H.Lév
Volume 18: 115–119 ELOPEA Publication date: 28 May 2015 T dx.doi.org/10.7751/telopea8510 Journal of Plant Systematics plantnet.rbgsyd.nsw.gov.au/Telopea • escholarship.usyd.edu.au/journals/index.php/TEL • ISSN 0312-9764 (Print) • ISSN 2200-4025 (Online) Lectotypification of Citrus cavaleriei H.Lév. ex Cavalerie (Rutaceae: Aurantioideae) David J. Mabberley1,2,3 and Phillip G. Kodela1 1National Herbarium of New South Wales, Royal Botanic Gardens & Domain Trust, Mrs Macquaries Road, Sydney, NSW 2000, Australia; 2Wadham College, University of Oxford, UK; 3Macquarie University, NSW 2109, Australia Abstract In the context of the elucidation of the ancestry of today’s commercial citrus crops, a lectotype is here designated for Citrus cavaleriei H.Lév. ex Cavalerie (Rutaceae, subfam. Aurantioideae), a species found in China and India, and one of the putative parents of C. ×junos Siebold ex Tanaka, the yuzu. Introduction The history of the domestication of citrus fruits is complicated by extensive hybridity between wild species, brought together by humans, and subsequent selection of apomictic clones, which make up the bulk of today’s commercial citrus crops worldwide (Mabberley 1997, 2004, 2013). The most significant globally are selections from the Citrus ×aurantium L. complex, oranges and grapefruit etc., derived from crosses between C. maxima (Burm.) Merr., the pomelo, and the mandarin from southern China, generally known as C. reticulata Blanco. The lemons and bergamots are derived from crosses between this complex and C. medica L., the citron. Even though this is well-established, there are still outstanding issues, in that no unequivocally wild populations of either C. -

Genomics of the Origin and Evolution of Citrus
OPEN ARTICLE doi:10.1038/nature25447 Genomics of the origin and evolution of Citrus Guohong Albert Wu1, Javier Terol2, Victoria Ibanez2, Antonio López-García2, Estela Pérez-Román2, Carles Borredá2, Concha Domingo2, Francisco R. Tadeo2, Jose Carbonell-Caballero3, Roberto Alonso3, Franck Curk4, Dongliang Du5, Patrick Ollitrault6, Mikeal L. Roose7, Joaquin Dopazo3,8, Frederick G. Gmitter Jr5, Daniel S. Rokhsar1,9,10 & Manuel Talon2 The genus Citrus, comprising some of the most widely cultivated fruit crops worldwide, includes an uncertain number of species. Here we describe ten natural citrus species, using genomic, phylogenetic and biogeographic analyses of 60 accessions representing diverse citrus germ plasms, and propose that citrus diversified during the late Miocene epoch through a rapid southeast Asian radiation that correlates with a marked weakening of the monsoons. A second radiation enabled by migration across the Wallace line gave rise to the Australian limes in the early Pliocene epoch. Further identification and analyses of hybrids and admixed genomes provides insights into the genealogy of major commercial cultivars of citrus. Among mandarins and sweet orange, we find an extensive network of relatedness that illuminates the domestication of these groups. Widespread pummelo admixture among these mandarins and its correlation with fruit size and acidity suggests a plausible role of pummelo introgression in the selection of palatable mandarins. This work provides a new evolutionary framework for the genus Citrus. The genus Citrus -

Citrus Genomes
March 31, 2021 [AgroScience Today] Popular Article Volume 2 Issue 3 Page: 0101 – 0103 Jagannadham Prasanth Tej Kumar Scientist (Biotechnology) ICAR Central Citrus Research Institute Amravati Road Nagpur Maharashtra India - 440 033 Citrus Genomes: Thirugnanavel Anbalagan Scientist (Fruit Science) Enigma Code Breaker ICAR Central Citrus Research Institute Amravati Road Nagpur Maharashtra India - 440 033 A genome is the complete set of instructions needed to build an Anjitha George organism and allow it to grow and develop. The genome sequence Scientist (Entomology) of any organism helps to understand its evolution and lays a ICAR Central Citrus Research Institute foundation for the functional characterization means for Amravati Road understanding the genetic basis of differences between plants and Nagpur other eukaryotes, and provides the foundation for detailed Maharashtra functional characterization of plant genes. The discovery of DNA India – 440 033 as a genetic material has revolutionized the science and researchers striving to decode the genetic information. The Kiran Kumar K arrival of next generation sequencing technologies has reduced Scientist (Nematology) the time and cost required to generate draft genomes. In citrus ICAR Central Citrus Research Institute group, 9 species were sequenced, several assemblies were Amravati Road available in public domain and several were in pipeline. The Nagpur analysis of genomes unraveled the origin, evolution of citrus Maharashtra species and identification genes involved in the characters typical India – 440 033 to citrus like apomixis, vitamin C synthesis. INTRODUCTION Corresponding Authors A genome harbours the information required for the functioning of an Jagannadham Prasanth Tej Kumar organisms. The scanning of genome can reveal information leading to [email protected] a particular phenotype. -

RUTACEAE 芸香科 Yun Xiang Ke Zhang Dianxiang (张奠湘)1; Thomas G
RUTACEAE 芸香科 yun xiang ke Zhang Dianxiang (张奠湘)1; Thomas G. Hartley2, David J. Mabberley3 Shrubs, trees, or sometimes herbs, sometimes scrambling or scandent, sometimes armed, with aromatic volatile oils contained in glands visible at surface of at least leaves, young branchlets, inflorescences, flower parts, fruit, or cotyledons in seed. Stipules absent [or stipular excrescences rarely present]. Leaves alternate, opposite [or whorled], simple (petiole neither apically swollen nor articulate with leaf blade), 1-foliolate (in individual specimens at least some 1-foliolate leaves with petiole apically swollen and/or articulate with leaf blade), or variously compound. Flowers bisexual or unisexual, usually 3–5-merous, actinomorphic or rarely zygomorphic, hypo- gynous [or rarely perigynous]. Perianth in 2 series, with clearly differentiated calyx and corolla or sometimes in 2 irregular series or 1 series, with ± undifferentiated tepals. Sepals distinct or connate to their full length. Petals distinct [or rarely coherent or connate for part of their length]. Stamens usually as many as or 2 × as many as petals or sometimes more numerous; filaments distinct or sometimes coherent or connate for at least part of their length; anthers introrse or sometimes latrorse, longitudinally dehiscent. Disk [rarely lack- ing] within androecium, nectariferous, flattened, annular, cup-shaped, pulvinate, or sometimes columnar, bell-shaped, conic, or hour- glass-shaped. Gynoecium of 1–5 distinct 1-loculed carpels or 2 to many partially to completely connate carpels; placentation axile [very rarely becoming parietal]; ovules 1 to many per locule. Fruit of 2–5 follicles [drupes or samaras] or a single follicle, capsule, or berry [or samara]. Seeds with relatively large embryo; endosperm present and fleshy or lacking. -

HUANGLONGBING and ITS VECTORS Information to Develop A
HUANGLONGBING AND ITS VECTORS Information to Develop a Response Plan for the Citrus and Allied Industries Patricia Barkley (formerly Citrus Australia Ltd. and NSW DPI) & GAC Beattie (University of Western Sydney) PATHOGENS COMMON NAMES huanglongbing (official common name, meaning ‘yellow shoot disease’), citrus greening (informal common name), likubin or decline (Taiwan), leaf mottling (Philippines), citrus dieback (India), citrus vein- phloem degeneration (Indonesia), citrus greening, yellow branch or blotchy-mottle (South Africa) SCIENTIFIC NAMES ‘Candidatus Liberibacter asiaticus’ (Asiatic form), ‘Ca. L. africanus’ 1(African form) and ‘Ca. L. americanus’ (‘South American’ form);2 SYNONYMS ‘Ca. Liberobacter asiaticus’ and ‘Ca. Liberobacter asiaticum’ ‘Ca. Liberobacter africanus’ and ‘Ca. Liberobacter africanum’ ASIATIC VECTOR COMMON NAMES Asiatic citrus psyllid, Asian citrus psyllid, oriental citrus psyllid, citrus psylla SCIENTIFIC NAME Diaphorina citri Kuwayama [Hemiptera: Sternorrhyncha: Psylloidea:Liviidae ] SYNONYMS Euphalerus citri Crawford AFRICAN VECTOR COMMON NAME African citrus psyllid, citrus psylla SCIENTIFIC NAME Trioza erytreae (del Guercio) [Hemiptera: Sternorrhyncha: Psylloidea: Triozidae] SYNONYMS Aleurodes erytreae del Guercio; Trioza citri Laing; Trioza merwei Petty; Spanioza merwei (Petty); Spanioza erythreae (del Guercio); Spanioza erytreae del Guercio 1 A sub-species transmitted by T. erytreae, ‘Ca. L. africanus subsp. capensis’, occurs in Cape chestnut (Calodendrum capense Thunb. [Rutaceae: Rutoideae]), an ornamental tree in southern Africa (Garnier et al. 2000, Pietersen et al. 2010). Pietersen & Viljoen (2012) and Viljoen et al (2013 a,b) found liberibacters in all genera of South African native Rutaceae analysed. Each rutaceous genus appears to harbour different specific Laf-like liberibacters. Those found in Xanthoxylum appear to be most closely related to Laf from Citrus. Vepris and Clausena harbour liberibacters more closely related to the LafC subspecies found in Calodendrum. -

Maladie Du Huanglongbing Analyse Du Risque Phytosanitaire Pour L’Union Européenne
Maladie du huanglongbing Analyse du risque phytosanitaire pour l’Union européenne Avis de l’Anses Rapport d’expertise collective Annexes et rapport annexe Avril 2019 - Édition scientifique Maladie du huanglongbing - Analyse du risque phytosanitaire pour l’Union européenne Maladie du huanglongbing Analyse du risque phytosanitaire pour l’Union européenne Avis de l’Anses Rapport d’expertise collective Annexes et rapport annexe Avril 2019 - Édition scientifique Avis de l’Anses Saisine n° « 2016-SA-0235 » Le directeur général Maisons-Alfort, le 25 avril 2019 AVIS de l’Agence nationale de sécurité sanitaire de l’alimentation, de l’environnement et du travail relatif à « une analyse de risque phytosanitaire pour la maladie du huanglongbing pour l’Union européenne » L’Anses met en œuvre une expertise scientifique indépendante et pluraliste. L’Anses contribue principalement à assurer la sécurité sanitaire dans les domaines de l’environnement, du travail et de l’alimentation et à évaluer les risques sanitaires qu’ils peuvent comporter. Elle contribue également à assurer d’une part la protection de la santé et du bien-être des animaux et de la santé des végétaux et d’autre part à l’évaluation des propriétés nutritionnelles des aliments. Elle fournit aux autorités compétentes toutes les informations sur ces risques ainsi que l’expertise et l’appui scientifique technique nécessaires à l’élaboration des dispositions législatives et réglementaires et à la mise en œuvre des mesures de gestion du risque (article L.1313-1 du code de la santé publique). Ses avis sont publiés sur son site internet. L’Anses a été saisie le 31 octobre 2016 par la DGAl pour la réalisation de l’expertise suivante : Saisine relative à une analyse de risque phytosanitaire pour la maladie du huanglongbing pour l’Union européenne. -

Ministério Da Agricultura, Pecuária E Abastecimento
Seção 1 ISSN 1677-7042 Nº 232, sexta-feira, 4 de dezembro de 2020 S EC R E T A R I A - G E R A L I - Ministro de Estado Chefe da Secretaria-Geral da Presidência da República, que o coordenará; PORTARIA Nº 98, DE 3 DE DEZEMBRO DE 2020 II - Secretário-Executivo; Aprova o Plano de Dados Abertos da Secretaria-Geral da Presidência da República -vigência 2020-2022. III - Secretário Especial de Modernização do Estado; O MINISTRO DE ESTADO CHEFE DA SECRETARIA-GERAL DA PRESIDÊNCIA DA IV - Secretário Especial de Administração; REPÚBLICA , no uso das atribuições que lhe conferem o artigo 87 da Constituição Federal, V - Secretário de Controle Interno; e considerando o que dispõem a Lei nº 12.527, de 18 de outubro de 2011, o Decreto nº 8.777, de 11 de maio de 2016, e a Resolução CGINDA nº 03, de 13 de outubro de 2017, VI - Subchefe para Assuntos Jurídicos; e que disciplinam a Política de Dados Abertos do Poder Executivo Federal, resolve: VII - Diretor-Geral da Imprensa Nacional. Art. 1º Aprovar o Plano de Dados Abertos da Secretaria-Geral da Presidência da § 1º Os membros de que tratam os incisos do caput serão representados, em República - vigência 2020-2022. suas ausências e impedimentos por seus substitutos no cargo em comissão ou função de confiança que ocupam. Art. 2º O referido Plano torna público o inventário de bases de dados do órgão,o § 2º O Gestor de Segurança da Informação da Secretaria-Geral da Presidência da resultado da consulta pública que identificou o interesse público pelas bases da SG/PR, a seleção República participará das reuniões do Comitê que tenham como pauta o tema da segurança dos dados que serão abertos, o cronograma de publicação, a descrição de ações de fomento ao da informação, coordenando, com direito a voto, a sessão específica referente ao tema.