Genomics of the Origin and Evolution of Citrus
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fruits; Fresh Vegetables and Fresh Limes” (Opp
Trademark Trial and Appeal Board Electronic Filing System. http://estta.uspto.gov ESTTA Tracking number: ESTTA881622 Filing date: 03/07/2018 IN THE UNITED STATES PATENT AND TRADEMARK OFFICE BEFORE THE TRADEMARK TRIAL AND APPEAL BOARD Proceeding 91238258 Party Plaintiff Wonderful Citrus LLC Correspondence DARYA P LAUFER ESQ Address ROLL LAW GROUP PC 11444 WEST OLYMPIC BLVD LOS ANGELES, CA 90064 UNITED STATES Email: [email protected], [email protected] Submission Other Motions/Papers Filer's Name Michael M. Vasseghi Filer's email [email protected], [email protected] Signature / Michael M. Vasseghi / Date 03/07/2018 Attachments Opposition with Exhibits-reduced size.pdf(1950576 bytes ) IN THE UNITED STATES PATENT AND TRADEMARK OFFICE TRADEMARK TRIAL AND APPEAL BOARD Wonderful Citrus LLC, Opposition No. 91238258 Opposer, Application Serial No. 87/472272 v. APB, Inc. dba Vision Produce Company, Applicant. OPPOSER WONDERFUL CITRUS LLC’S OPPOSITION TO APPLICANT’S MOTION FOR JUDGMENT ON THE PLEADINGS I. INTRODUCTION Applicant moves for judgment on the pleadings (“Motion”), arguing that “there is no genuine issue as to Opposer’s lack of prior rights in a trademark that could be confusingly similar to Applicant’s Mark.” (Motion pg. 3.)1 Applicant’s Motion is not well taken. It acknowledges that Opposer has alleged exactly what it takes issue with – that Opposer has prior rights in a trademark that could be confusingly similar to Applicant’s Mark. Despite this, Applicant seeks to take issue with those allegations, implicitly contending that Opposer will be unable to prove what it has alleged. (Motion pg. 2.) This is not a proper basis for judgment on the pleadings, which must accept as true all allegations asserted in the Opposition. -

Genetic Diversity and Population Structure of Pummelo (Citrus Maxima) Germplasm in China
Tree Genetics & Genomes (2017) 13: 58 DOI 10.1007/s11295-017-1133-0 ORIGINAL ARTICLE Genetic diversity and population structure of pummelo (Citrus maxima) germplasm in China Huiwen Yu1 & Xiaoming Yang 1 & Fei Guo1 & Xiaolin Jiang1 & Xiuxin Deng1 & Qiang Xu1 Received: 31 July 2016 /Revised: 11 March 2017 /Accepted: 19 March 2017 /Published online: 26 April 2017 # Springer-Verlag Berlin Heidelberg 2017 Abstract Pummelo (Citrus maxima) is one of the basic spe- Keywords Pummelo . Genetic diversity . Population cies of Citrus. It has been cultivated for about 4000 years in structure . Nuclear simple sequence repeat (nSSR) China, and therefore, there are abundant germplasm during the long time of culture. However, there is still a lack of a detailed study of the genetic characteristics of pummelo pop- Introduction ulation. In this study, genetic diversity and population struc- ture among 274 pummelo accessions collected in China were Citrus is one of the most important fruit crops in the world. analyzed using 31 nuclear simple sequence repeat (nSSR) The genetic background of citrus is very complicated because markers. The observed heterozygosity was calculated as of its biological characteristics, such as wide sexual compati- 0.325 and genetic differentiation Fst as 0.077. Genetic struc- bility on interspecies and intergenus levels. The complex ge- ture analysis divided the whole germplasm into three subpop- netic background has hindered the genetic studies in citrus. ulations, Pop-a, Pop-b, and Pop-c. Pop-a was composed of Exploring genetic variation within a single species will facil- accessions mostly from Southeast China, Pop-b was com- itate genetic analysis such as genome-wide association studies posed of accessions from the central region of South China, (GWAS) of important traits. -
Spicy Grapefruit Margarita. the RECIPE
THE RECIPE spicy grapefruit margarita. By halfbakedharvest I swear I have found the perfect amount of tequila, to lime juice, to fruity mixer for margs prep time 10 minutes total time 10 minutes servings 6 drinks 4.47 from 39 votes calories 207 kcal INGREDIENTS BIG BATCH MARGARITAS 1 1/2 cups silver tequila 12 ounces 2 cups grapefruit juice 16 ounces 3/4 cups fresh lime juice 1/4 cup agave nectar may also use honey 1-2 whole jalapeños halved (depending on how spicy you want your drink) slices lime + grapefruit for serving TO MAKE JUST ONE DRINK 1 1/2 - 2 ounces silver tequila 2 1/2 ounces grapefruit juice 1 ounce fresh lime juice 1 tablespoon agave nectar or more to taste 1-2 slices jalapeno depending on how spicy you want your drink slices lime + grapefruit for serving SPICY CHILI SALT RIM 1 tablespoon chili powder 1 tablespoon kosher salt 2 teaspoons granulated sugar INSTRUCTIONS SPICY CHILI SALT RIM In a bowl, combine the chili power salt and sugar. Mix well to combine. Use this mixture to rim your glasses. BIG BATCH MARGARITA Combine all the ingredients in a pitcher and stir to combine. Keep chilled in the fridge for 1-3 hours. Taste the drink every hour to check for spiciness. The longer you leave the jalapeño in the drink the spicier the drink will become. Once the drink is spiced to your liking, remove the jalapeños. When ready to serve, salt the rims of 4-6 glasses. Add ice to each glass and pour the margaritas over the ice. -

Reaction of Tangerines Genotypes to Elsinoe Fawcettiiunder
Reaction of tangerines genotypes to Elsinoe fawcettii under natural infection conditions Crop Breeding and Applied Biotechnology 11: 77-81, 2011 Brazilian Society of Plant Breeding. Printed in Brazil Reaction of tangerines genotypes to Elsinoe fawcettii under natural infection conditions Marcelo Claro de Souza1*, Eduardo Sanches Stuchi2 and Antonio de Goes3 Received 11 February 2010 Accepted 30 September 2010 ABSTRACT - A citrus scab disease, caused by Elsinoe fawcettii, is currently found in all citrus areas throughout Brazil. That being, given the importance of this casual agent, the behavior of tangerines and hybrids influenced by this pathogen was evaluated under natural infection conditions. This study was performed with plants around 15 years old without irrigation; 100 fruits of three plants were collected during harvest season, using a grade scale varying from 0 (absence of symptoms) to 6 (severe symptoms) the level of disease severity was determined. Among the cultivars, citrus scab resistance was observed in Citrus deliciosa, C. tangerina, C. nobilis; a mandarin hybrid (C. nobilis x C. deliciosa) and a satsuma hybrid (C. unshiu x C. sinensis). Among the other genotypes, symptoms were observed with levels of severity ranging from 1 to 3, indicating moderate resistance. Key words: Citrus scab, citrus crop, resistant varieties. INTRODUCTION In Brazil, E. fawcettii is responsible for citrus scab. The disease is widespread in many humid, citrus-cultivating In many citrus production areas around the world, areas around the world and decreases fruit values on the Elsinoe fawcettii is one of the main fungi diseases found. fresh-fruit market (Feichtenberger et al. 1986). In young It attacks a wide variety of citrus species and cultivars, plants or under severe infection, it may cause significant resulting in scab disease on leaves, twigs, and fruits (Timmer fruit drop. -

"Performance of Citrus Scion Cultivars and Rootstocks in a High-Density
REPORTS HORTSCIENCE 26(7):837-840. 1991. house and planted in the field in 1981. A split plot experiment and analysis of variance Performance of Citrus Scion Cultivars (ANOVA) statistics were used with four rep- lications, with cultivar as the main plot and and Rootstock in a High-density rootstock as the subplot. Field plots were four ´ four trees, with data taken from the Planting center four of the 16 trees. They were planted 1.5 m in the row and 3.3 m between rows T.A. Wheaton, W.S. Castle, J.D. Whitney, and D.P.H. Tucker and were irrigated and fertigated as required Citrus Research and Education Center, University of Florida, Institute of to maintain optimal soil water and nutrient levels using one microsprinkler per two trees. Food and Agricultural Sciences, 700 Experiment Station Road, Lake Trees were mechanically hedged and topped Alfred, FL 33850 during Summer 1987 and hedged again in 1989 to maintain a 1.5-m alley between rows Additional index words. tree spacing, yield efficiency and a 2.5-m tree height. Thus, the canopy Abstract. ‘Hamlin’ and ‘Valencia’ oranges [Citrus sinensis (L.) Osb.], ‘Murcott’ tangor size allocated for each tree was 1.5 m in the (C. reticulata Blanco ´ C. sinensis), and ‘Redblush’ grapefruit (C. paradisi Macf.) on row, 1.8 m across the row, and 2.5 m in 15 rootstock and own-rooted cuttings were planted at a 1.5 ´ 3.3-m spacing providing height, providing 6.8 m3 of canopy volume a density of 2020 trees/ha. -

Bright Citron™ Soak
Product Profile BRIGHT CITRON™ SOAK WHAT IT IS An aromatic and refreshing sea salt soak for cleansing skin. WHAT IT DOES Softens and cleanses skin. WHY YOU NEED IT • Provides the first step to cleanse impurities and soften skin for a luxurious manicure or pedicure experience. • Gently cleanses without drying skin. FRAGRANCE FEATURED Pink Grapefruit and Warm Amber. FRAGRANCE PROFILE • Bright citrus top notes. • Soft floral mid notes. CREATIVE SUGGESTIONS • Light woody base notes. • Create a fragrant ceremonial experience by placing the Soak into warm water directly prior to ACTIVE BOTANICALS immersing hands or feet. Kaffir Lime (Citrus Hystrix Leaf Extract): • Add lime or grapefruit slices to enhance the Is known to purify skin and promote radiance. experience. • Use as a foot soak prior to all salon services. Honey: • Excellent retail product for home use in bath tubs. Is known to hydrate skin and help retain moisture. INGREDIENT LISTING FEATURED INGREDIENTS & BENEFITS Maris Sal ((Sea Salt) Sel Marin), Sodium Sesquicarbonate, Sea Salt: Argania Spinosa Kernel Oil, Honey, Citrus Hystrix Leaf Natural salt used for its therapeutic properties. Extract, Aqua ((Water) Eau), Glycerin, Propylene Glycol, Salts increase the flow of water in and out of Hydrated Silica, Parfum (Fragrance), Limonene, Linalool, cells, in essence flushing and cleansing the cells. Hexyl Cinnamal, Citral. This facilitates the absorption of other ingredients into the cells. AVAILABLE SIZES • Retail/Professional: 410 g (14.4 oz) Argan Oil (Argania Spinosa Kernel Oil): • Professional: 3.3 kg (118.8 oz) Is known to nourish dry skin. DIRECTIONS FOR USE • For feet, add a teaspoon to footbath and agitate water to dissolve. -

Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid
Known Host Plants of Huanglongbing (HLB) and Asian Citrus Psyllid Diaphorina Liberibacter citri Plant Name asiaticus Citrus Huanglongbing Psyllid Aegle marmelos (L.) Corr. Serr.: bael, Bengal quince, golden apple, bela, milva X Aeglopsis chevalieri Swingle: Chevalier’s aeglopsis X X Afraegle gabonensis (Swingle) Engl.: Gabon powder-flask X Afraegle paniculata (Schum.) Engl.: Nigerian powder- flask X Atalantia missionis (Wall. ex Wight) Oliv.: see Pamburus missionis X X Atalantia monophylla (L.) Corr.: Indian atalantia X Balsamocitrus dawei Stapf: Uganda powder- flask X X Burkillanthus malaccensis (Ridl.) Swingle: Malay ghost-lime X Calodendrum capense Thunb.: Cape chestnut X × Citroncirus webberi J. Ingram & H. E. Moore: citrange X Citropsis gilletiana Swingle & M. Kellerman: Gillet’s cherry-orange X Citropsis schweinfurthii (Engl.) Swingle & Kellerm.: African cherry- orange X Citrus amblycarpa (Hassk.) Ochse: djerook leemo, djeruk-limau X Citrus aurantiifolia (Christm.) Swingle: lime, Key lime, Persian lime, lima, limón agrio, limón ceutí, lima mejicana, limero X X Citrus aurantium L.: sour orange, Seville orange, bigarde, marmalade orange, naranja agria, naranja amarga X Citrus depressa Hayata: shiikuwasha, shekwasha, sequasse X Citrus grandis (L.) Osbeck: see Citrus maxima X Citrus hassaku hort. ex Tanaka: hassaku orange X Citrus hystrix DC.: Mauritius papeda, Kaffir lime X X Citrus ichangensis Swingle: Ichang papeda X Citrus jambhiri Lushington: rough lemon, jambhiri-orange, limón rugoso, rugoso X X Citrus junos Sieb. ex Tanaka: xiang -

Lime (Citrus Aurantifolia (Christm.) Swingle) Essential Oils: Volatile Compounds, Antioxidant Capacity, and Hypolipidemic Effect
foods Article Lime (Citrus aurantifolia (Christm.) Swingle) Essential Oils: Volatile Compounds, Antioxidant Capacity, and Hypolipidemic Effect Li-Yun Lin 1,*, Cheng-Hung Chuang 2 , Hsin-Chun Chen 3 and Kai-Min Yang 4 1 Department of Food Science and Technology, Hungkuang University, Taichung 433, Taiwan 2 Department of Nutrition, Hungkuang University, Taichung 433, Taiwan 3 Department of Cosmeceutics, China Medical University, Taichung 404, Taiwan 4 Department of Hospitality Management, Mingdao Unicersity, ChangHua 523, Taiwan * Correspondence: [email protected]; Tel.: +886-33-432-793 Received: 19 July 2019; Accepted: 2 September 2019; Published: 7 September 2019 Abstract: Lime peels are mainly obtained from the byproducts of the juice manufacturing industry, which we obtained and used to extract essential oil (2.3%) in order to examine the antioxidant and hypolipidaemic effects. We identified 60 volatile compounds of lime essential oil (LEO) with GC/MS, of which the predominant constituents were limonene, γ-terpinene, and β-pinene. Lime essential oil was measured according to the DPPH assay and ABTS assay, with IC50 values of 2.36 mg/mL and 0.26 mg/mL, respectively. This study also explored the protective effects of LEO against lipid-induced hyperlipidemia in a rat model. Two groups of rats received oral LEO in doses of 0.74 g/100 g and 2.23 g/100 g with their diets. Eight weeks later, we found that the administration of LEO improved the serum total cholesterol, triglyceride, low-density lipoprotein cholesterol, alanine aminotransferase, and aspartate transaminase levels in the hyperlipidemic rats (p < 0.05). Simultaneously, the LEO improved the health of the rats in terms of obesity, atherogenic index, and fatty liver. -

Tropical Horticulture: Lecture 32 1
Tropical Horticulture: Lecture 32 Lecture 32 Citrus Citrus: Citrus spp., Rutaceae Citrus are subtropical, evergreen plants originating in southeast Asia and the Malay archipelago but the precise origins are obscure. There are about 1600 species in the subfamily Aurantioideae. The tribe Citreae has 13 genera, most of which are graft and cross compatible with the genus Citrus. There are some tropical species (pomelo). All Citrus combined are the most important fruit crop next to grape. 1 Tropical Horticulture: Lecture 32 The common features are a superior ovary on a raised disc, transparent (pellucid) dots on leaves, and the presence of aromatic oils in leaves and fruits. Citrus has increased in importance in the United States with the development of frozen concentrate which is much superior to canned citrus juice. Per-capita consumption in the US is extremely high. Citrus mitis (calamondin), a miniature orange, is widely grown as an ornamental house pot plant. History Citrus is first mentioned in Chinese literature in 2200 BCE. First citrus in Europe seems to have been the citron, a fruit which has religious significance in Jewish festivals. Mentioned in 310 BCE by Theophrastus. Lemons and limes and sour orange may have been mutations of the citron. The Romans grew sour orange and lemons in 50–100 CE; the first mention of sweet orange in Europe was made in 1400. Columbus brought citrus on his second voyage in 1493 and the first plantation started in Haiti. In 1565 the first citrus was brought to the US in Saint Augustine. 2 Tropical Horticulture: Lecture 32 Taxonomy Citrus classification based on morphology of mature fruit (e.g. -

Classification and Cultivars
1 Classification and Cultivars 2 Two Tribes • Clauseneae • Citreae has 3 Subtribes –Triphasiinae –Balsamocitrineae –Citrinae 3 Fortunella • Four species - Small trees and shrubs. • Flowers later than Citrus. • Freeze - hardy • Small fruit –‘Meiwa’ and ‘Marumi’ - round –‘Nagami’ ovate 4 Poncirus • Two trifoliate spp. –trifoliata ‘Flying Dragon’ –poyandra • Deciduous • Thorny, Cold hardy, long thorns • Makes great hedges , rootstocks 5 Microcitrus • Northeastern rainforest Australia • Moderate-sized trees. • Leaves are unifoliate dimorphic • Microcitrus australasica –Resistant to burrowing nematode and phytophthora • Micro leaves, flowers, and fruit 6 Clymenia • Unifoliate acuminate leaves tapering into very short petiole. • Branches are thornless. • Style shorter than other true Citrus and stigma is larger and flattened • Fruit - ovoid, thin peeled, many oil glands, many small seeds. 7 Eremocitrus • Xerophytic native of Australia • Spreading long drooping branches • Leaves unifoliate, greyish green, thick, leatherly, and lanceolate. • Sunken stomata, freeze hardy • Ideal xeroscape plant. 8 Citrus - Subgenus Eucitrus • Vesicles - no acrid or bitter oil • C. medica (Citrons) –Uses - candied peel, • Jewish ceremony • Exocortis indicator 9 Citrus limon (Lemons) • Commerce –‘Lisbon’ and ‘Eureka’ • Dooryard –Meyer (Lemon hybrid) • Rough Lemon –Rootstock 10 Lemon Hybrids • Lemonage (lemon x sweet orange) • Lemonime (lemon x lime) • Lemandrin (lemon x mandarin) • Eremolemon (Eremocitrus x lemon) - Australian Desert Lemon 11 Citrus aurantifolia (Limes) • ‘Key’ or ‘Mexican’ limes • ‘Tahiti’ or ‘Persian’ limes some are triploids and seedless • C. macrophylla (lime-like fruit) –Rootstock in California • Lemonimes (lime x lemon) • Limequats (lime x kumquat) 12 • Not grown either in Tahiti or Persian (Iran) • Seedless and marketed when still dark green 13 C. aurantium - Sour Orange • ‘Seville’ in Southern Europe –Orange marmalade • ‘Bouquet’ & ‘Bergamot’ • - Italy –Essential oil • Many forms like ‘Bittersweet’ –Rootstock - High quality fruit. -

Chemical Variability of Peel and Leaf Essential Oils in the Citrus Subgenus Papeda (Swingle) and Relatives
Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives Clémentine Baccati, Marc Gibernau, Mathieu Paoli, Patrick Ollitrault, Félix Tomi, François Luro To cite this version: Clémentine Baccati, Marc Gibernau, Mathieu Paoli, Patrick Ollitrault, Félix Tomi, et al.. Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives. Plants, MDPI, 2021, 10 (6), pp.1117. 10.3390/plants10061117. hal-03262123 HAL Id: hal-03262123 https://hal.archives-ouvertes.fr/hal-03262123 Submitted on 16 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives Clémentine Baccati 1, Marc Gibernau 1, Mathieu Paoli 1, Patrick Ollitrault 2,3, Félix Tomi 1, * and François Luro 2 1 Université de Corse-CNRS, UMR 6134 SPE, Route des Sanguinaires, 20000 Ajaccio, France; [email protected] (C.B.) ; [email protected] (M.G.) ; [email protected] (M.P.) ; [email protected] (F.T.) 2 UMR AGAP Institut, Univ Montpellier, CIRAD, INRAE, Institut Agro – 20230, San Giuliano, France 3 CIRAD, UMR AGAP, F-20230 San Giuliano, France * Correspondence: [email protected]; tel.:+33-495-52-4122. -
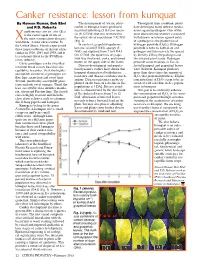
Canker Resistance: Lesson from Kumquat by Naveen Kumar, Bob Ebel the Development of Asiatic Citrus Throughout Their Evolution, Plants and P.D
Canker resistance: lesson from kumquat By Naveen Kumar, Bob Ebel The development of Asiatic citrus Throughout their evolution, plants and P.D. Roberts canker in kumquat leaves produced have developed many defense mecha- anthomonas citri pv. citri (Xcc) localized yellowing (5 DAI) or necro- nisms against pathogens. One of the is the causal agent of one of sis (9-12 DAI) that was restricted to most characteristic features associated the most serious citrus diseases the actual site of inoculation 7-12 DAI with disease resistance against entry X (Fig. 2). of a pathogen is the production of worldwide, Asiatic citrus canker. In the United States, Florida experienced In contrast, grapefruit epidermis hydrogen peroxide (H2O2). Hydrogen three major outbreaks of Asiatic citrus became raised (5 DAI), spongy (5 peroxide is toxic to both plant and canker in 1910, 1984 and 1995, and it DAI) and ruptured from 7 to 8 DAI. pathogen and thus restricts the spread is a constant threat to the $9 billion On 12 DAI, the epidermis of grape- by directly killing the pathogen and citrus industry. fruit was thickened, corky, and turned the infected plant tissue. Hydrogen Citrus genotypes can be classified brown on the upper side of the leaves. peroxide concentrations in Xcc-in- into four broad classes based on sus- Disease development and popula- fected kumquat and grapefruit leaves ceptibility to canker. First, the highly- tion dynamics studies have shown that were different. Kumquat produces susceptible commercial genotypes are kumquat demonstrated both disease more than three times the amount of Key lime, grapefruit and sweet lime.