Whole Genome Association Analysis of Treatment Response in Obsessive-Compulsive Disorder
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
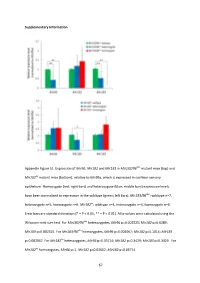
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

Transport of Sugars
BI84CH32-Frommer ARI 29 April 2015 12:34 Transport of Sugars Li-Qing Chen,1,∗ Lily S. Cheung,1,∗ Liang Feng,3 Widmar Tanner,2 and Wolf B. Frommer1 1Department of Plant Biology, Carnegie Institution for Science, Stanford, California 94305; email: [email protected] 2Zellbiologie und Pflanzenbiochemie, Universitat¨ Regensburg, 93040 Regensburg, Germany 3Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, California 94305 Annu. Rev. Biochem. 2015. 84:865–94 Keywords First published online as a Review in Advance on glucose, sucrose, carrier, GLUT, SGLT, SWEET March 5, 2015 The Annual Review of Biochemistry is online at Abstract biochem.annualreviews.org Soluble sugars serve five main purposes in multicellular organisms: as sources This article’s doi: of carbon skeletons, osmolytes, signals, and transient energy storage and as 10.1146/annurev-biochem-060614-033904 transport molecules. Most sugars are derived from photosynthetic organ- Copyright c 2015 by Annual Reviews. isms, particularly plants. In multicellular organisms, some cells specialize All rights reserved in providing sugars to other cells (e.g., intestinal and liver cells in animals, ∗ These authors contributed equally to this review. photosynthetic cells in plants), whereas others depend completely on an ex- Annu. Rev. Biochem. 2015.84:865-894. Downloaded from www.annualreviews.org ternal supply (e.g., brain cells, roots and seeds). This cellular exchange of Access provided by b-on: Universidade de Lisboa (UL) on 09/05/16. For personal use only. sugars requires transport proteins to mediate uptake or release from cells or subcellular compartments. Thus, not surprisingly, sugar transport is criti- cal for plants, animals, and humans. -

Supplemental Table 3 Site ID Intron Poly(A) Site Type NM/KG Inum
Supplemental Table 3 Site ID Intron Poly(A) site Type NM/KG Inum Region Gene ID Gene Symbol Gene Annotation Hs.120277.1.10 chr3:170997234:170996860 170996950 b NM_153353 7 CDS 151827 LRRC34 leucine rich repeat containing 34 Hs.134470.1.27 chr17:53059664:53084458 53065543 b NM_138962 10 CDS 124540 MSI2 musashi homolog 2 (Drosophila) Hs.162889.1.18 chr14:80367239:80329208 80366262 b NM_152446 12 CDS 145508 C14orf145 chromosome 14 open reading frame 145 Hs.187898.1.27 chr22:28403623:28415294 28404458 b NM_181832 16 3UTR 4771 NF2 neurofibromin 2 (bilateral acoustic neuroma) Hs.228320.1.6 chr10:115527009:115530350 115527470 b BC036365 5 CDS 79949 C10orf81 chromosome 10 open reading frame 81 Hs.266308.1.2 chr11:117279579:117278191 117278967 b NM_032046 12 CDS 84000 TMPRSS13 transmembrane protease, serine 13 Hs.266308.1.4 chr11:117284536:117281662 117283722 b NM_032046 9 CDS 84000 TMPRSS13 transmembrane protease, serine 13 Hs.2689.1.4 chr10:53492398:53563605 53492622 b NM_006258 7 CDS 5592 PRKG1 protein kinase, cGMP-dependent, type I Hs.280781.1.6 chr18:64715646:64829150 64715837 b NM_024781 4 CDS 79839 C18orf14 chromosome 18 open reading frame 14 Hs.305985.2.25 chr12:8983686:8984438 8983942 b BX640639 17 3UTR NA NA NA Hs.312098.1.36 chr1:151843991:151844258 151844232 b NM_003815 15 CDS 8751 ADAM15 a disintegrin and metalloproteinase domain 15 (metargidin) Hs.314338.1.11 chr21:39490293:39481214 39487623 b NM_018963 41 CDS 54014 BRWD1 bromodomain and WD repeat domain containing 1 Hs.33368.1.3 chr15:92685158:92689361 92688314 b NM_018349 6 CDS 55784 MCTP2 multiple C2-domains with two transmembrane regions 2 Hs.346736.1.21 chr2:99270738:99281614 99272414 b AK126402 10 3UTR 51263 MRPL30 mitochondrial ribosomal protein L30 Hs.445061.1.19 chr16:69322898:69290216 69322712 b NM_018052 14 CDS 55697 VAC14 Vac14 homolog (S. -

Strand Breaks for P53 Exon 6 and 8 Among Different Time Course of Folate Depletion Or Repletion in the Rectosigmoid Mucosa
SUPPLEMENTAL FIGURE COLON p53 EXONIC STRAND BREAKS DURING FOLATE DEPLETION-REPLETION INTERVENTION Supplemental Figure Legend Strand breaks for p53 exon 6 and 8 among different time course of folate depletion or repletion in the rectosigmoid mucosa. The input of DNA was controlled by GAPDH. The data is shown as ΔCt after normalized to GAPDH. The higher ΔCt the more strand breaks. The P value is shown in the figure. SUPPLEMENT S1 Genes that were significantly UPREGULATED after folate intervention (by unadjusted paired t-test), list is sorted by P value Gene Symbol Nucleotide P VALUE Description OLFM4 NM_006418 0.0000 Homo sapiens differentially expressed in hematopoietic lineages (GW112) mRNA. FMR1NB NM_152578 0.0000 Homo sapiens hypothetical protein FLJ25736 (FLJ25736) mRNA. IFI6 NM_002038 0.0001 Homo sapiens interferon alpha-inducible protein (clone IFI-6-16) (G1P3) transcript variant 1 mRNA. Homo sapiens UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 GALNTL5 NM_145292 0.0001 (GALNT15) mRNA. STIM2 NM_020860 0.0001 Homo sapiens stromal interaction molecule 2 (STIM2) mRNA. ZNF645 NM_152577 0.0002 Homo sapiens hypothetical protein FLJ25735 (FLJ25735) mRNA. ATP12A NM_001676 0.0002 Homo sapiens ATPase H+/K+ transporting nongastric alpha polypeptide (ATP12A) mRNA. U1SNRNPBP NM_007020 0.0003 Homo sapiens U1-snRNP binding protein homolog (U1SNRNPBP) transcript variant 1 mRNA. RNF125 NM_017831 0.0004 Homo sapiens ring finger protein 125 (RNF125) mRNA. FMNL1 NM_005892 0.0004 Homo sapiens formin-like (FMNL) mRNA. ISG15 NM_005101 0.0005 Homo sapiens interferon alpha-inducible protein (clone IFI-15K) (G1P2) mRNA. SLC6A14 NM_007231 0.0005 Homo sapiens solute carrier family 6 (neurotransmitter transporter) member 14 (SLC6A14) mRNA. -

Glucose Transporters As a Target for Anticancer Therapy
cancers Review Glucose Transporters as a Target for Anticancer Therapy Monika Pliszka and Leszek Szablewski * Chair and Department of General Biology and Parasitology, Medical University of Warsaw, 5 Chalubinskiego Str., 02-004 Warsaw, Poland; [email protected] * Correspondence: [email protected]; Tel.: +48-22-621-26-07 Simple Summary: For mammalian cells, glucose is a major source of energy. In the presence of oxygen, a complete breakdown of glucose generates 36 molecules of ATP from one molecule of glucose. Hypoxia is a hallmark of cancer; therefore, cancer cells prefer the process of glycolysis, which generates only two molecules of ATP from one molecule of glucose, and cancer cells need more molecules of glucose in comparison with normal cells. Increased uptake of glucose by cancer cells is due to increased expression of glucose transporters. However, overexpression of glucose transporters, promoting the process of carcinogenesis, and increasing aggressiveness and invasiveness of tumors, may have also a beneficial effect. For example, upregulation of glucose transporters is used in diagnostic techniques such as FDG-PET. Therapeutic inhibition of glucose transporters may be a method of treatment of cancer patients. On the other hand, upregulation of glucose transporters, which are used in radioiodine therapy, can help patients with cancers. Abstract: Tumor growth causes cancer cells to become hypoxic. A hypoxic condition is a hallmark of cancer. Metabolism of cancer cells differs from metabolism of normal cells. Cancer cells prefer the process of glycolysis as a source of ATP. Process of glycolysis generates only two molecules of ATP per one molecule of glucose, whereas the complete oxidative breakdown of one molecule of glucose yields 36 molecules of ATP. -

Clinical, Molecular and Genetic Aspects
Gaceta Médica de México. 2016;152 Contents available at PubMed www.anmm.org.mx PERMANYER Gac Med Mex. 2016;152:492-501 www.permanyer.com GACETA MÉDICA DE MÉXICO REVIEW ARTICLE Glucotransporters: clinical, molecular and genetic aspects Roberto de Jesús Sandoval-Muñiz, Belinda Vargas-Guerrero, Luis Javier Flores-Alvarado and Carmen Magdalena Gurrola-Díaz* Health Sciences Campus, University of Guadalajara, Guadalajara, Jal., Mexico Abstract Oxidation of glucose is the major source of obtaining cell energy, this process requires glucose transport into the cell. However, cell membranes are not permeable to polar molecules such as glucose; therefore its internalization is accomplished by transporter proteins coupled to the cell membrane. In eukaryotic cells, there are two types of carriers coupled to the membrane: 1) cotransporter Na+-glucose (SGLT) where Na+ ion provides motive power for the glucose´s internalization, and 2) the glucotransporters (GLUT) act by facilitated diffusion. This review will focus on the 14 GLUT so far described. Despite the structural homology of GLUT, different genetic alterations of each GLUT cause specific clinical entities. Therefore, the aim of this review is to gather the molecular and biochemical available information of each GLUT as well as the particular syndromes and pathologies related with GLUT´s alterations and their clinical approaches. (Gac Med Mex. 2016;152:492-501) Corresponding author: Carmen Magdalena Gurrola-Díaz, [email protected] KEY WORDS: Sugar transport facilitators. GLUT. Glucose transporters. SLC2A. different affinity for carbohydrates1. In eukaryote cells ntroduction I there are two membrane-coupled transporter proteins: 1) Sodium-glucose co-transporters (SGLT), located in Glucose metabolism provides energy to the cell by the small bowel and renal tissue, mainly responsible means of adenosine-5’-triphosphate (ATP) biosynthe- for the absorption and reabsorption of nutrients, and sis, with glycolysis as the catabolic pathway. -

Table SI. Enriched Genes in the Upregulated Genes of the Recovery Group According to the GO Molecular Function Terms. A, Downreg
Table SI. Enriched genes in the upregulated genes of the recovery group according to the GO Molecular Function terms. A, Downregulated genes Adjusted Total Molecular Rank P‑value genes (n) Function Genes 1 <0.001 266 GO:0019899 Raf1 Timp1 Tbc1d8 Ube2g2 Ube2z enzyme binding Lonrf3 Tbc1d15 Rnf144a Ube2g1 Shc3 Rgcc Rnf19a Ube2j2 Rnf138 Atg13 Cks1b Ube2j1 Rnf19b Trib1 Trib3 Abtb2 Rnf125 Cdc42ep3 Nploc4 Cdc42ep4 Cdc42ep2 Rab11fip5 Arih2 Brms1 Tmem189 Mef2d Hspb1 Cdk9 Ksr1 Tnfaip3 Net1 Rnf180 Fgr Bhlhe41 Irs2 Ppp1r15a Asb4 Trim72 Zfp36 Sfn Xpo6 Fap Sox9 Mapk7 Itga3 Tubb5 Daxx Klf4 Stat3 Gab2 Myo9b Cstb Hmox1 Por Bcl2l1 Plin5 Chp1 Ube2i Sash1 Sqstm1 Rxra Slpi Sdc4 Tnfaip1 Cd40 Slc12a4 Map2k3 Ywhah Ppp1r12a Cry1 Plek Egfr Tnip1 Npc1l1 Rock2 Map2k6 Per1 Nfkbia Bdkrb2 Prkch Hif1a Golga5 Ripk1 Map3k1 Glud1 Nufip1 Clu Spry2 Hcls1 Ifnar2 Tuba1b Cdkn1a Sik1 Tmem173 Map3k2 Tnf Riok3 Ptpn2 Cep192 Smad2 Fas Jak2 Ankrd1 Rela Rps6ka4 Ankrd2 Rabgef1 Prkar1b Nop58 Casp8 Cflar Hdac4 Sele Nek2 Optn Nek6 Lcn2 Stom Traf6 Spred1 Nop56 Src Ccnl1 Ptpn22 Il6ra Pip5k1a F3 Bcl10 3110043O21Rik Tnfrsf1b Slc2a1 Sfpq Rpa2 Errfi1 Mad2l2 Tbc1d14 Uchl1 Glmn Scarb2 Ulk1 Ung Rad18 Mef2a Ctsc Ipo5 Mvp Kctd13 Msn Eif4ebp1 Casp3 Smad1 Ubash3b Ets1 Tirap Smad3 Tgfbr2 Ptgs2 Prr5l Micall1 Cnppd1 Map2k4 Tnks1bp1 Ppp1r32 Prdm4 Midn Ibtk Rusc2 Fmnl2 Ptpn23 Sh3bp4 Nop14 Kdm1a Serpine1 Gch1 Inf2 Csf3 Snx10 Txnip Egr1 Ranbp9 Akap12 Rab3gap2 Ddx58 Bcor Rabggta Pik3r1 Pkp2 Usp22 Shc1 Ptpn11 Fzd5 Cxcr4 Plaur Bag5 Maml1 Camk2n2 Taf7 Ywhag Ezr Jun Camk2d Parp4 Nod2 Ptafr Hmga2 Zfp746 Ptk2b Flot1 -

The Concise Guide to Pharmacology 2019/20: Transporters
University of Dundee The Concise Guide to Pharmacology 2019/20 CGTP Collaborators; Alexander, Stephen P. H.; Kelly, Eamonn; Mathie, Alistair; Peters, John A.; Veale, Emma L. Published in: British Journal of Pharmacology DOI: 10.1111/bph.14753 Publication date: 2019 Document Version Publisher's PDF, also known as Version of record Link to publication in Discovery Research Portal Citation for published version (APA): CGTP Collaborators, Alexander, S. P. H., Kelly, E., Mathie, A., Peters, J. A., Veale, E. L., ... Davies, J. A. (2019). The Concise Guide to Pharmacology 2019/20: Transporters. British Journal of Pharmacology, 176 (S1), S397- S493. https://doi.org/10.1111/bph.14753 General rights Copyright and moral rights for the publications made accessible in Discovery Research Portal are retained by the authors and/or other copyright owners and it is a condition of accessing publications that users recognise and abide by the legal requirements associated with these rights. • Users may download and print one copy of any publication from Discovery Research Portal for the purpose of private study or research. • You may not further distribute the material or use it for any profit-making activity or commercial gain. • You may freely distribute the URL identifying the publication in the public portal. Take down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Download date: 07. Dec. 2019 S.P.H. Alexander et al. The Concise -

RNA-Seq Reveals Conservation of Function Among the Yolk Sacs Of
RNA-seq reveals conservation of function among the PNAS PLUS yolk sacs of human, mouse, and chicken Tereza Cindrova-Daviesa, Eric Jauniauxb, Michael G. Elliota,c, Sungsam Gongd,e, Graham J. Burtona,1, and D. Stephen Charnock-Jonesa,d,e,1,2 aCentre for Trophoblast Research, Department of Physiology, Development and Neuroscience, University of Cambridge, Cambridge, CB2 3EG, United Kingdom; bElizabeth Garret Anderson Institute for Women’s Health, Faculty of Population Health Sciences, University College London, London, WC1E 6BT, United Kingdom; cSt. John’s College, University of Cambridge, Cambridge, CB2 1TP, United Kingdom; dDepartment of Obstetrics and Gynaecology, University of Cambridge, Cambridge, CB2 0SW, United Kingdom; and eNational Institute for Health Research, Cambridge Comprehensive Biomedical Research Centre, Cambridge, CB2 0QQ, United Kingdom Edited by R. Michael Roberts, University of Missouri-Columbia, Columbia, MO, and approved May 5, 2017 (received for review February 14, 2017) The yolk sac is phylogenetically the oldest of the extraembryonic yolk sac plays a critical role during organogenesis (3–5, 8–10), membranes. The human embryo retains a yolk sac, which goes there are limited data to support this claim. Obtaining experi- through primary and secondary phases of development, but its mental data for the human is impossible for ethical reasons, and importance is controversial. Although it is known to synthesize thus we adopted an alternative strategy. Here, we report RNA proteins, its transport functions are widely considered vestigial. sequencing (RNA-seq) data derived from human and murine yolk Here, we report RNA-sequencing (RNA-seq) data for the human sacs and compare them with published data from the yolk sac of and murine yolk sacs and compare those data with data for the the chicken. -

Electronic Supplementary Material (ESI) for Molecular Omics
Electronic Supplementary Material (ESI) for Molecular Omics. This journal is © The Royal Society of Chemistry 2020 SUPPLEMENTARY INFORMATION Supplementary Table S2. GO analysis for the 289 overlapping genes based on DAVID database. ID Category Term Genes PValue HSD3B2, CYP24A1, ME3, SORD, ADHFE1, CYP2C44, PAH, HIBADH, MTHFD1, ALDH1A1, PECR, CYP4A12A, FMO1, MIOX, CYP2J11, HAAO, BC089597, ALDH4A1, BDH2, DAO, NQO1, BDH1, SARDH, HPD, 1 BP GO:0055114~oxidation reduction 1.18E-17 NOX4, GCDH, SUOX, AKR1E1, QDPR, CMAH, HGD, FADS2, AKR1C21, PPARGC1A, TET1, DDO, NNT, HAO2, CYP2D26, HSD11B2, DIO1, RDH16, CYP4A14, STEAP1, DCXR, PRODH SLC12A6, SLC2A13, SLC5A2, SLC12A1, SLC5A1, SLC22A7, SLC22A8, RHBG, SLC7A9, AQP6, SLC26A4, GO:0055085~transmembrane 2 BP SLC23A1, SLC16A7, SLC2A4, RHCG, SLC7A1, 1.28E-07 transport SLC25A10, SLC2A2, SLC16A9, SLC5A9, SLC13A2, SLC25A37, SLC13A3, SLC46A1, SLC5A12 PDK2, SLC37A4, PDK4, CMAH, PGAM2, ADIPOQ, GO:0005996~monosaccharide 3 BP HIBADH, PCK1, GALM, G6PC, PPP1R1A, GYS2, MYC, 1.25E-06 metabolic process DCXR, XYLB SLC12A6, SLC5A2, SLC23A1, SLC12A1, SLC5A1, 4 BP GO:0006814~sodium ion transport SLC9A3, SLC5A9, SLC13A2, SLC13A3, SLC10A2, 3.05E-06 SLC4A4, SLC5A12 SLC5A2, G6PC, SLC2A4, SLC2A2, SLC5A1, SLC37A4, 5 BP GO:0015758~glucose transport 3.22E-06 STXBP4 SLC5A2, G6PC, SLC2A4, SLC2A2, SLC5A1, SLC37A4, 6 BP GO:0008645~hexose transport 4.95E-06 STXBP4 GO:0015749~monosaccharide SLC5A2, G6PC, SLC2A4, SLC2A2, SLC5A1, SLC37A4, 7 BP 6.06E-06 transport STXBP4 GO:0006006~glucose metabolic PDK2, G6PC, PPP1R1A, PDK4, SLC37A4, GYS2, -

Vast Human-Specific Delay in Cortical Ontogenesis Associated With
Supplementary information Extension of cortical synaptic development distinguishes humans from chimpanzees and macaques Supplementary Methods Sample collection We used prefrontal cortex (PFC) and cerebellar cortex (CBC) samples from postmortem brains of 33 human (aged 0-98 years), 14 chimpanzee (aged 0-44 years) and 44 rhesus macaque individuals (aged 0-28 years) (Table S1). Human samples were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland, USA, the Netherlands Brain Bank, Amsterdam, Netherlands and the Chinese Brain Bank Center, Wuhan, China. Informed consent for use of human tissues for research was obtained in writing from all donors or their next of kin. All subjects were defined as normal by forensic pathologists at the corresponding brain bank. All subjects suffered sudden death with no prolonged agonal state. Chimpanzee samples were obtained from the Yerkes Primate Center, GA, USA, the Anthropological Institute & Museum of the University of Zürich-Irchel, Switzerland and the Biomedical Primate Research Centre, Netherlands (eight Western chimpanzees, one Central/Eastern and five of unknown origin). Rhesus macaque samples were obtained from the Suzhou Experimental Animal Center, China. All non-human primates used in this study suffered sudden deaths for reasons other than their participation in this study and without any relation to the tissue used. CBC dissections were made from the cerebellar cortex. PFC dissections were made from the frontal part of the superior frontal gyrus. All samples contained an approximately 2:1 grey matter to white matter volume ratio. RNA microarray hybridization RNA isolation, hybridization to microarrays, and data preprocessing were performed as described previously (Khaitovich et al.