Electronic Supplementary Material (ESI) for Molecular Omics
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

Remodeling Adipose Tissue Through in Silico Modulation of Fat Storage For
Chénard et al. BMC Systems Biology (2017) 11:60 DOI 10.1186/s12918-017-0438-9 RESEARCHARTICLE Open Access Remodeling adipose tissue through in silico modulation of fat storage for the prevention of type 2 diabetes Thierry Chénard2, Frédéric Guénard3, Marie-Claude Vohl3,4, André Carpentier5, André Tchernof4,6 and Rafael J. Najmanovich1* Abstract Background: Type 2 diabetes is one of the leading non-infectious diseases worldwide and closely relates to excess adipose tissue accumulation as seen in obesity. Specifically, hypertrophic expansion of adipose tissues is related to increased cardiometabolic risk leading to type 2 diabetes. Studying mechanisms underlying adipocyte hypertrophy could lead to the identification of potential targets for the treatment of these conditions. Results: We present iTC1390adip, a highly curated metabolic network of the human adipocyte presenting various improvements over the previously published iAdipocytes1809. iTC1390adip contains 1390 genes, 4519 reactions and 3664 metabolites. We validated the network obtaining 92.6% accuracy by comparing experimental gene essentiality in various cell lines to our predictions of biomass production. Using flux balance analysis under various test conditions, we predict the effect of gene deletion on both lipid droplet and biomass production, resulting in the identification of 27 genes that could reduce adipocyte hypertrophy. We also used expression data from visceral and subcutaneous adipose tissues to compare the effect of single gene deletions between adipocytes from each -

Small Cell Ovarian Carcinoma: Genomic Stability and Responsiveness to Therapeutics
Gamwell et al. Orphanet Journal of Rare Diseases 2013, 8:33 http://www.ojrd.com/content/8/1/33 RESEARCH Open Access Small cell ovarian carcinoma: genomic stability and responsiveness to therapeutics Lisa F Gamwell1,2, Karen Gambaro3, Maria Merziotis2, Colleen Crane2, Suzanna L Arcand4, Valerie Bourada1,2, Christopher Davis2, Jeremy A Squire6, David G Huntsman7,8, Patricia N Tonin3,4,5 and Barbara C Vanderhyden1,2* Abstract Background: The biology of small cell ovarian carcinoma of the hypercalcemic type (SCCOHT), which is a rare and aggressive form of ovarian cancer, is poorly understood. Tumourigenicity, in vitro growth characteristics, genetic and genomic anomalies, and sensitivity to standard and novel chemotherapeutic treatments were investigated in the unique SCCOHT cell line, BIN-67, to provide further insight in the biology of this rare type of ovarian cancer. Method: The tumourigenic potential of BIN-67 cells was determined and the tumours formed in a xenograft model was compared to human SCCOHT. DNA sequencing, spectral karyotyping and high density SNP array analysis was performed. The sensitivity of the BIN-67 cells to standard chemotherapeutic agents and to vesicular stomatitis virus (VSV) and the JX-594 vaccinia virus was tested. Results: BIN-67 cells were capable of forming spheroids in hanging drop cultures. When xenografted into immunodeficient mice, BIN-67 cells developed into tumours that reflected the hypercalcemia and histology of human SCCOHT, notably intense expression of WT-1 and vimentin, and lack of expression of inhibin. Somatic mutations in TP53 and the most common activating mutations in KRAS and BRAF were not found in BIN-67 cells by DNA sequencing. -

Viewed Under 23 (B) Or 203 (C) fi M M Male Cko Mice, and Largely Unaffected Magni Cation; Scale Bars, 500 M (B) and 50 M (C)
BRIEF COMMUNICATION www.jasn.org Renal Fanconi Syndrome and Hypophosphatemic Rickets in the Absence of Xenotropic and Polytropic Retroviral Receptor in the Nephron Camille Ansermet,* Matthias B. Moor,* Gabriel Centeno,* Muriel Auberson,* † † ‡ Dorothy Zhang Hu, Roland Baron, Svetlana Nikolaeva,* Barbara Haenzi,* | Natalya Katanaeva,* Ivan Gautschi,* Vladimir Katanaev,*§ Samuel Rotman, Robert Koesters,¶ †† Laurent Schild,* Sylvain Pradervand,** Olivier Bonny,* and Dmitri Firsov* BRIEF COMMUNICATION *Department of Pharmacology and Toxicology and **Genomic Technologies Facility, University of Lausanne, Lausanne, Switzerland; †Department of Oral Medicine, Infection, and Immunity, Harvard School of Dental Medicine, Boston, Massachusetts; ‡Institute of Evolutionary Physiology and Biochemistry, St. Petersburg, Russia; §School of Biomedicine, Far Eastern Federal University, Vladivostok, Russia; |Services of Pathology and ††Nephrology, Department of Medicine, University Hospital of Lausanne, Lausanne, Switzerland; and ¶Université Pierre et Marie Curie, Paris, France ABSTRACT Tight control of extracellular and intracellular inorganic phosphate (Pi) levels is crit- leaves.4 Most recently, Legati et al. have ical to most biochemical and physiologic processes. Urinary Pi is freely filtered at the shown an association between genetic kidney glomerulus and is reabsorbed in the renal tubule by the action of the apical polymorphisms in Xpr1 and primary fa- sodium-dependent phosphate transporters, NaPi-IIa/NaPi-IIc/Pit2. However, the milial brain calcification disorder.5 How- molecular identity of the protein(s) participating in the basolateral Pi efflux remains ever, the role of XPR1 in the maintenance unknown. Evidence has suggested that xenotropic and polytropic retroviral recep- of Pi homeostasis remains unknown. Here, tor 1 (XPR1) might be involved in this process. Here, we show that conditional in- we addressed this issue in mice deficient for activation of Xpr1 in the renal tubule in mice resulted in impaired renal Pi Xpr1 in the nephron. -

Upregulation of Peroxisome Proliferator-Activated Receptor-Α And
Upregulation of peroxisome proliferator-activated receptor-α and the lipid metabolism pathway promotes carcinogenesis of ampullary cancer Chih-Yang Wang, Ying-Jui Chao, Yi-Ling Chen, Tzu-Wen Wang, Nam Nhut Phan, Hui-Ping Hsu, Yan-Shen Shan, Ming-Derg Lai 1 Supplementary Table 1. Demographics and clinical outcomes of five patients with ampullary cancer Time of Tumor Time to Age Differentia survival/ Sex Staging size Morphology Recurrence recurrence Condition (years) tion expired (cm) (months) (months) T2N0, 51 F 211 Polypoid Unknown No -- Survived 193 stage Ib T2N0, 2.41.5 58 F Mixed Good Yes 14 Expired 17 stage Ib 0.6 T3N0, 4.53.5 68 M Polypoid Good No -- Survived 162 stage IIA 1.2 T3N0, 66 M 110.8 Ulcerative Good Yes 64 Expired 227 stage IIA T3N0, 60 M 21.81 Mixed Moderate Yes 5.6 Expired 16.7 stage IIA 2 Supplementary Table 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of an ampullary cancer microarray using the Database for Annotation, Visualization and Integrated Discovery (DAVID). This table contains only pathways with p values that ranged 0.0001~0.05. KEGG Pathway p value Genes Pentose and 1.50E-04 UGT1A6, CRYL1, UGT1A8, AKR1B1, UGT2B11, UGT2A3, glucuronate UGT2B10, UGT2B7, XYLB interconversions Drug metabolism 1.63E-04 CYP3A4, XDH, UGT1A6, CYP3A5, CES2, CYP3A7, UGT1A8, NAT2, UGT2B11, DPYD, UGT2A3, UGT2B10, UGT2B7 Maturity-onset 2.43E-04 HNF1A, HNF4A, SLC2A2, PKLR, NEUROD1, HNF4G, diabetes of the PDX1, NR5A2, NKX2-2 young Starch and sucrose 6.03E-04 GBA3, UGT1A6, G6PC, UGT1A8, ENPP3, MGAM, SI, metabolism -

Rhbg and Rhcg, the Putative Ammonia Transporters, Are Expressed in the Same Cells in the Distal Nephron
ARTICLES J Am Soc Nephrol 14: 545–554, 2003 RhBG and RhCG, the Putative Ammonia Transporters, Are Expressed in the Same Cells in the Distal Nephron FABIENNE QUENTIN,* DOMINIQUE ELADARI,*† LYDIE CHEVAL,‡ CLAUDE LOPEZ,§ DOMINIQUE GOOSSENS,§ YVES COLIN,§ JEAN-PIERRE CARTRON,§ MICHEL PAILLARD,*† and RE´ GINE CHAMBREY* *Institut National de la Sante´et de la Recherche Me´dicale Unite´356, Institut Fe´de´ratif de Recherche 58, Universite´Pierre et Marie Curie, Paris, France; †Hoˆpital Europe´en Georges Pompidou, Assistance Publique- Hoˆpitaux de Paris, Paris, France; ‡Centre National de la Recherche Scientifique FRE 2468, Paris, France; and §Institut National de la Sante´et de la Recherche Me´dicale Unite´76, Institut National de la Transfusion Sanguine, Paris, France. Abstract. Two nonerythroid homologs of the blood group Rh RhBG expression in distal nephron segments within the cortical proteins, RhCG and RhBG, which share homologies with specific labyrinth, medullary rays, and outer and inner medulla. RhBG ammonia transporters in primitive organisms and plants, could expression was restricted to the basolateral membrane of epithelial represent members of a new family of proteins involved in am- cells. The same localization was observed in rat and mouse monia transport in the mammalian kidney. Consistent with this kidney. RT-PCR analysis on microdissected rat nephron segments hypothesis, the expression of RhCG was recently reported at the confirmed that RhBG mRNAs were chiefly expressed in CNT apical pole of all connecting tubule (CNT) cells as well as in and cortical and outer medullary CD. Double immunostaining intercalated cells of collecting duct (CD). To assess the localiza- with RhCG demonstrated that RhBG and RhCG were coex- tion along the nephron of RhBG, polyclonal antibodies against the pressed in the same cells, but with a basolateral and apical local- Rh type B glycoprotein were generated. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -
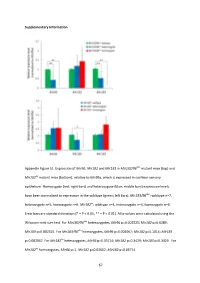
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

Taqman® Openarray® Pharmacogenomics (Pgx) Panel
PRODUCT OVERVIEW TaqMan® OpenArray® Pharmacogenomics (PGx) Panel TaqMan® OpenArray® Pharmacogenomics (PGx) Panel Genotyping analysis of drug metabolism enzymes and associated transport proteins The TaqMan® OpenArray® Trusted TaqMan® performance samples, studies, and labs. Assays Pharmacogenomics (PGx) Panel is Each TaqMan® DME Genotyping were selected for optimal relevance a powerful tool to help researchers Assay contains two allele-specific to current pharmacogenomics study human genetic variation probes and a primer pair to detect studies and organized for a simplified in relation to drug action and its the specific SNP target. Both the workflow. potential application to medical probes and primers uniquely align ® ® treatment. within the genome, enabling the The TaqMan OpenArray PGx TaqMan® genotyping technology to Panel provides valuable data for ® ® The TaqMan OpenArray PGx provide superior specificity. It is this the study of drug interactions in Panel was developed for quick and specificity that allows these assays several research areas. Targeted easy screening of known high- to detect targets residing in highly genes relate to areas of study such value target genes associated with homologous gene families that may as cardiovascular (CYP2D6, CYP2C19, drug metabolism enzymes and include pseudogenes. TaqMan® Drug NAT1, NAT2), analgesics (CYP2C9, associated transport proteins. Metabolism Enzyme Genotyping CYP2D6), rheumatology (CYP2C9, The panel consists of 158 drug Assays were developed using a high TPMT), neurology (CYP2C19, CYP2D6), metabolism enzyme (DME) assays level of bioinformatics and wet-lab and musculoskeletal (CYP2C19). A derived from the PharmaADME stringency. All assays have passed total of 29 genes are covered across Core Marker Set (Table 1). The performance tests involving 180 158 unique assays. -

The Role of the Renal Ammonia Transporter Rhcg in Metabolic Responses to Dietary Protein
BASIC RESEARCH www.jasn.org The Role of the Renal Ammonia Transporter Rhcg in Metabolic Responses to Dietary Protein † † † Lisa Bounoure,* Davide Ruffoni, Ralph Müller, Gisela Anna Kuhn, Soline Bourgeois,* Olivier Devuyst,* and Carsten A. Wagner* *Institute of Physiology and Zurich Center for Integrative Human Physiology, University of Zurich, Zurich, Switzerland; and †Institute for Biomechanics, ETH Zurich, Zurich, Switzerland ABSTRACT High dietary protein imposes a metabolic acid load requiring excretion and buffering by the kidney. Impaired acid excretion in CKD, with potential metabolic acidosis, may contribute to the progression of CKD. Here, we investigated the renal adaptive response of acid excretory pathways in mice to high- protein diets containing normal or low amounts of acid-producing sulfur amino acids (SAA) and examined how this adaption requires the RhCG ammonia transporter. Diets rich in SAA stimulated expression of + enzymes and transporters involved in mediating NH4 reabsorption in the thick ascending limb of the loop of Henle. The SAA-rich diet increased diuresis paralleled by downregulation of aquaporin-2 (AQP2) water + channels. The absence of Rhcg transiently reduced NH4 excretion, stimulated the ammoniagenic path- 2 way more strongly, and further enhanced diuresis by exacerbating the downregulation of the Na+/K+/2Cl cotransporter (NKCC2) and AQP2, with less phosphorylation of AQP2 at serine 256. The high protein acid load affected bone turnover, as indicated by higher Ca2+ and deoxypyridinoline excretion, phenomena exaggerated in the absence of Rhcg. In animals receiving a high-protein diet with low SAA content, the + kidney excreted alkaline urine, with low levels of NH4 and no change in bone metabolism. -

Screening of Genetic Variations of SLC15A2, SLC22A1, SLC22A2 and SLC22A6 Genes
Journal of Human Genetics (2011) 56, 666–670 & 2011 The Japan Society of Human Genetics All rights reserved 1434-5161/11 $32.00 www.nature.com/jhg ORIGINAL ARTICLE Screening of genetic variations of SLC15A2, SLC22A1, SLC22A2 and SLC22A6 genes Hyun Sub Cheong1,4, Hae Deun Kim2,4, Han Sung Na2,JiOnKim1, Lyoung Hyo Kim1, Seung Hee Kim2, Joon Seol Bae3, Myeon Woo Chung2 and Hyoung Doo Shin1,3 A growing list of membrane-spanning proteins involved in the transport of a large variety of drugs has been recognized and characterized to include peptide and organic anion/cation transporters. Given such an important role of transporter genes in drug disposition process, the role of single-nucleotide polymorphisms (SNPs) in such transporters as potential determinants of interindividual variability in drug disposition and pharmacological response has been investigated. To define the distribution of transporter gene SNPs across ethnic groups, we screened 450 DNAs in cohorts of 250 Korean, 50 Han Chinese, 50 Japanese, 50 African-American and 50 European-American ancestries for 64 SNPs in four transporter genes encoding proteins of the solute carrier family (SLC15A2, SLC22A1, SLC22A2 and SLC22A6). Of the 64 SNPs, 19 were core pharmacogenetic variants and 45 were HapMap tagging SNPs. Polymorphisms were genotyped using the golden gate genotyping assay. After genetic variability, haplotype structures and ethnic diversity were analyzed, we observed that the distributions of SNPs in a Korean population were similar to other Asian groups (Chinese and Japanese), and significantly different from African-American and European-American cohorts. Findings from this study would be valuable for further researches, including pharmacogenetic studies for drug responses.