Supplemental Table 3 Site ID Intron Poly(A) Site Type NM/KG Inum
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Information
doi: 10.1038/nature08795 SUPPLEMENTARY INFORMATION Supplementary Discussion Population naming In some contexts, the indigenous hunter-gatherer and pastoralist peoples of southern Africa are referred to collectively as the Khoisan (Khoi-San) or more recently Khoesan (Khoe-San) people. This grouping is based on the unique linguistic use of click-consonants1. Many names, often country-specific, have been used by Bantu pastoralists and European settlers to describe the hunter-gatherers, including San, Saan, Sonqua, Soaqua, Souqua, Sanqua, Kwankhala, Basarwa, Batwa, Abathwa, Baroa, Bushmen, Bossiesmans, Bosjemans, or Bosquimanos. In addition, group-specific names such as !Kung and Khwe are often used for the broader population. The two most commonly used names, “San” and “Bushmen”, have both been associated with much controversy due to derogatory connotations2. “San” has become the more popular term used in Western literature, although “Bushmen” is arguably the more commonly recognized term within the communities. Since they have no collective name for themselves, the term Bushmen was selected for use in this paper as the term most familiar to the participants themselves. Regarding identification of individuals The five men identified in this study have all elected to have their identity made public knowledge. Thus we present two complete personal genomes (KB1 and ABT), a low-coverage personal genome (NB1), and personal exomes for all five men. On a scientific level, identification allows for current and future correlation of genetic data with demographic and medical histories. On a social level, identification allows for maximizing community benefit. For !Gubi, G/aq’o, D#kgao and !Aî, their name represents not only themselves, but importantly their extended family unit and a way of life severely under threat. -

The Switch from Fermentation to Respiration in Saccharomyces Cerevisiae Is Regulated by the Ert1 Transcriptional Activator/Repressor
INVESTIGATION The Switch from Fermentation to Respiration in Saccharomyces cerevisiae Is Regulated by the Ert1 Transcriptional Activator/Repressor Najla Gasmi,* Pierre-Etienne Jacques,† Natalia Klimova,† Xiao Guo,§ Alessandra Ricciardi,§ François Robert,†,** and Bernard Turcotte*,‡,§,1 ‡Department of Medicine, *Department of Biochemistry, and §Department of Microbiology and Immunology, McGill University Health Centre, McGill University, Montreal, QC, Canada H3A 1A1, †Institut de recherches cliniques de Montréal, Montréal, QC, Canada H2W 1R7, and **Département de Médecine, Faculté de Médecine, Université de Montréal, QC, Canada H3C 3J7 ABSTRACT In the yeast Saccharomyces cerevisiae, fermentation is the major pathway for energy production, even under aerobic conditions. However, when glucose becomes scarce, ethanol produced during fermentation is used as a carbon source, requiring a shift to respiration. This adaptation results in massive reprogramming of gene expression. Increased expression of genes for gluconeogenesis and the glyoxylate cycle is observed upon a shift to ethanol and, conversely, expression of some fermentation genes is reduced. The zinc cluster proteins Cat8, Sip4, and Rds2, as well as Adr1, have been shown to mediate this reprogramming of gene expression. In this study, we have characterized the gene YBR239C encoding a putative zinc cluster protein and it was named ERT1 (ethanol regulated transcription factor 1). ChIP-chip analysis showed that Ert1 binds to a limited number of targets in the presence of glucose. The strongest enrichment was observed at the promoter of PCK1 encoding an important gluconeogenic enzyme. With ethanol as the carbon source, enrichment was observed with many additional genes involved in gluconeogenesis and mitochondrial function. Use of lacZ reporters and quantitative RT-PCR analyses demonstrated that Ert1 regulates expression of its target genes in a manner that is highly redundant with other regulators of gluconeogenesis. -

Binnenwerk Cindy Postma.Indd
CHAPTER 6 Multiple putative oncogenes at the chromosome 20q amplicon contribute to colorectal adenoma to carcinoma progression Gut 2009, 58: 79-89 Beatriz Carvalho Cindy Postma Sandra Mongera Erik Hopmans Sharon Diskin Mark A. van de Wiel Wim van Criekinge Olivier Thas Anja Matthäi Miguel A. Cuesta Jochim S. Terhaar sive Droste Mike Craanen Evelin Schröck Bauke Ylstra Gerrit A. Meijer 104 | Chapter 6 Abstract Objective: This study aimed to identify the oncogenes at 20q involved in colorectal adenoma to carcinoma progression by measuring the effect of 20q gain on mRNA expression of genes in this amplicon. Methods: Segmentation of DNA copy number changes on 20q was performed by array CGH in 34 non-progressed colorectal adenomas, 41 progressed adenomas (i.e. adenomas that present a focus of cancer) and 33 adenocarcinomas. Moreover, a robust analysis of altered expression of genes in these segments was performed by microarray analysis in 37 adenomas and 31 adenocarcinomas. Protein expression was evaluated by immunohistochemistry on tissue microarrays. Results: The genes C20orf24, AURKA, RNPC1, TH1L, ADRM1, C20orf20 and TCFL5, mapping at 20q were signifi cantly overexpressed in carcinomas compared to adenomas as consequence of copy number gain of 20q. Conclusion: This approach revealed C20orf24, AURKA, RNPC1, TH1L, ADRM1, C20orf20 and TCFL5 genes to be important in chromosomal instability-related adenoma to carcinoma progression. These genes therefore may serve as highly specifi c biomarkers for colorectal cancer with potential clinical applications. Putative oncogenes at chromosome 20q in colorectal carcinogenesis | 105 Introduction The majority of cancers are epithelial in origin and arise through a stepwise progression from normal cells, through dysplasia, into malignant cells that invade surrounding tissues and have metastatic potential. -

Figure S1. Representative Report Generated by the Ion Torrent System Server for Each of the KCC71 Panel Analysis and Pcafusion Analysis
Figure S1. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. A Figure S1. Continued. Representative report generated by the Ion Torrent system server for each of the KCC71 panel analysis and PCaFusion analysis. (A) Details of the run summary report followed by the alignment summary report for the KCC71 panel analysis sequencing. (B) Details of the run summary report for the PCaFusion panel analysis. B Figure S2. Comparative analysis of the variant frequency found by the KCC71 panel and calculated from publicly available cBioPortal datasets. For each of the 71 genes in the KCC71 panel, the frequency of variants was calculated as the variant number found in the examined cases. Datasets marked with different colors and sample numbers of prostate cancer are presented in the upper right. *Significantly high in the present study. Figure S3. Seven subnetworks extracted from each of seven public prostate cancer gene networks in TCNG (Table SVI). Blue dots represent genes that include initial seed genes (parent nodes), and parent‑child and child‑grandchild genes in the network. Graphical representation of node‑to‑node associations and subnetwork structures that differed among and were unique to each of the seven subnetworks. TCNG, The Cancer Network Galaxy. Figure S4. REVIGO tree map showing the predicted biological processes of prostate cancer in the Japanese. Each rectangle represents a biological function in terms of a Gene Ontology (GO) term, with the size adjusted to represent the P‑value of the GO term in the underlying GO term database. -

The Concise Guide to Pharmacology 2019/20
Edinburgh Research Explorer THE CONCISE GUIDE TO PHARMACOLOGY 2019/20 Citation for published version: Cgtp Collaborators 2019, 'THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters', British Journal of Pharmacology, vol. 176 Suppl 1, pp. S397-S493. https://doi.org/10.1111/bph.14753 Digital Object Identifier (DOI): 10.1111/bph.14753 Link: Link to publication record in Edinburgh Research Explorer Document Version: Publisher's PDF, also known as Version of record Published In: British Journal of Pharmacology General rights Copyright for the publications made accessible via the Edinburgh Research Explorer is retained by the author(s) and / or other copyright owners and it is a condition of accessing these publications that users recognise and abide by the legal requirements associated with these rights. Take down policy The University of Edinburgh has made every reasonable effort to ensure that Edinburgh Research Explorer content complies with UK legislation. If you believe that the public display of this file breaches copyright please contact [email protected] providing details, and we will remove access to the work immediately and investigate your claim. Download date: 28. Sep. 2021 S.P.H. Alexander et al. The Concise Guide to PHARMACOLOGY 2019/20: Transporters. British Journal of Pharmacology (2019) 176, S397–S493 THE CONCISE GUIDE TO PHARMACOLOGY 2019/20: Transporters Stephen PH Alexander1 , Eamonn Kelly2, Alistair Mathie3 ,JohnAPeters4 , Emma L Veale3 , Jane F Armstrong5 , Elena Faccenda5 ,SimonDHarding5 ,AdamJPawson5 , Joanna L -
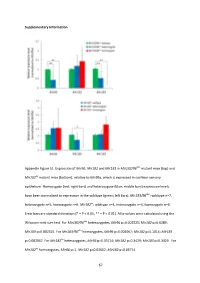
And Mir183 in Mir183/96 Dko Mutant Mice (Top) And
Supplementary Information Appendix Figure S1. Expression of Mir96 , Mir182 and Mir183 in Mir183/96 dko mutant mice (top) and Mir182 ko mutant mice (bottom), relative to Mir99a , which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96 dko : wildtype n=7, heterozygote n=5, homozygote n=6. Mir182 ko : wildtype n=4, heterozygote n=4, homozygote n=4. Error bars are standard deviation (* = P < 0.05, ** = P < 0.01). All p-values were calculated using the Wilcoxon rank sum test. For Mir183/96 dko heterozygotes, Mir96 p=0.002525; Mir182 p=0.6389; Mir183 p=0.002525. For Mir183/96 dko homozygotes, Mir96 p=0.002067; Mir182 p=0.1014; Mir183 p=0.002067. For Mir182 ko heterozygotes, Mir96 p=0.05714; Mir182 p=0.3429; Mir183 p=0.3429. For Mir182 ko homozygotes, Mir96 p=1; Mir182 p=0.02652; Mir183 p=0.05714. 67 68 Appendix Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96 dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 69 70 Appendix Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182 ko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. 71 Appendix Figure S4. Mean ABR waveforms at 12kHz, shown at 20dB (top) and 50dB (bottom) above threshold (sensation level, SL) ± standard deviation, at four weeks old. -

The Interactome of KRAB Zinc Finger Proteins Reveals the Evolutionary History of Their Functional Diversification
Resource The interactome of KRAB zinc finger proteins reveals the evolutionary history of their functional diversification Pierre-Yves Helleboid1,†, Moritz Heusel2,†, Julien Duc1, Cécile Piot1, Christian W Thorball1, Andrea Coluccio1, Julien Pontis1, Michaël Imbeault1, Priscilla Turelli1, Ruedi Aebersold2,3,* & Didier Trono1,** Abstract years ago (MYA) (Imbeault et al, 2017). Their products harbor an N-terminal KRAB (Kru¨ppel-associated box) domain related to that of Krüppel-associated box (KRAB)-containing zinc finger proteins Meisetz (a.k.a. PRDM9), a protein that originated prior to the diver- (KZFPs) are encoded in the hundreds by the genomes of higher gence of chordates and echinoderms, and a C-terminal array of zinc vertebrates, and many act with the heterochromatin-inducing fingers (ZNF) with sequence-specific DNA-binding potential (Urru- KAP1 as repressors of transposable elements (TEs) during early tia, 2003; Birtle & Ponting, 2006; Imbeault et al, 2017). KZFP genes embryogenesis. Yet, their widespread expression in adult tissues multiplied by gene and segment duplication to count today more and enrichment at other genetic loci indicate additional roles. than 350 and 700 representatives in the human and mouse Here, we characterized the protein interactome of 101 of the ~350 genomes, respectively (Urrutia, 2003; Kauzlaric et al, 2017). A human KZFPs. Consistent with their targeting of TEs, most KZFPs majority of human KZFPs including all primate-restricted family conserved up to placental mammals essentially recruit KAP1 and members target sequences derived from TEs, that is, DNA trans- associated effectors. In contrast, a subset of more ancient KZFPs posons, ERVs (endogenous retroviruses), LINEs, SINEs (long and rather interacts with factors related to functions such as genome short interspersed nuclear elements, respectively), or SVAs (SINE- architecture or RNA processing. -

Table SI. Genes Upregulated ≥ 2-Fold by MIH 2.4Bl Treatment Affymetrix ID
Table SI. Genes upregulated 2-fold by MIH 2.4Bl treatment Fold UniGene ID Description Affymetrix ID Entrez Gene Change 1558048_x_at 28.84 Hs.551290 231597_x_at 17.02 Hs.720692 238825_at 10.19 93953 Hs.135167 acidic repeat containing (ACRC) 203821_at 9.82 1839 Hs.799 heparin binding EGF like growth factor (HBEGF) 1559509_at 9.41 Hs.656636 202957_at 9.06 3059 Hs.14601 hematopoietic cell-specific Lyn substrate 1 (HCLS1) 202388_at 8.11 5997 Hs.78944 regulator of G-protein signaling 2 (RGS2) 213649_at 7.9 6432 Hs.309090 serine and arginine rich splicing factor 7 (SRSF7) 228262_at 7.83 256714 Hs.127951 MAP7 domain containing 2 (MAP7D2) 38037_at 7.75 1839 Hs.799 heparin binding EGF like growth factor (HBEGF) 224549_x_at 7.6 202672_s_at 7.53 467 Hs.460 activating transcription factor 3 (ATF3) 243581_at 6.94 Hs.659284 239203_at 6.9 286006 Hs.396189 leucine rich single-pass membrane protein 1 (LSMEM1) 210800_at 6.7 1678 translocase of inner mitochondrial membrane 8 homolog A (yeast) (TIMM8A) 238956_at 6.48 1943 Hs.741510 ephrin A2 (EFNA2) 242918_at 6.22 4678 Hs.319334 nuclear autoantigenic sperm protein (NASP) 224254_x_at 6.06 243509_at 6 236832_at 5.89 221442 Hs.374076 adenylate cyclase 10, soluble pseudogene 1 (ADCY10P1) 234562_x_at 5.89 Hs.675414 214093_s_at 5.88 8880 Hs.567380; far upstream element binding protein 1 (FUBP1) Hs.707742 223774_at 5.59 677825 Hs.632377 small nucleolar RNA, H/ACA box 44 (SNORA44) 234723_x_at 5.48 Hs.677287 226419_s_at 5.41 6426 Hs.710026; serine and arginine rich splicing factor 1 (SRSF1) Hs.744140 228967_at 5.37 -

Transport of Sugars
BI84CH32-Frommer ARI 29 April 2015 12:34 Transport of Sugars Li-Qing Chen,1,∗ Lily S. Cheung,1,∗ Liang Feng,3 Widmar Tanner,2 and Wolf B. Frommer1 1Department of Plant Biology, Carnegie Institution for Science, Stanford, California 94305; email: [email protected] 2Zellbiologie und Pflanzenbiochemie, Universitat¨ Regensburg, 93040 Regensburg, Germany 3Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, California 94305 Annu. Rev. Biochem. 2015. 84:865–94 Keywords First published online as a Review in Advance on glucose, sucrose, carrier, GLUT, SGLT, SWEET March 5, 2015 The Annual Review of Biochemistry is online at Abstract biochem.annualreviews.org Soluble sugars serve five main purposes in multicellular organisms: as sources This article’s doi: of carbon skeletons, osmolytes, signals, and transient energy storage and as 10.1146/annurev-biochem-060614-033904 transport molecules. Most sugars are derived from photosynthetic organ- Copyright c 2015 by Annual Reviews. isms, particularly plants. In multicellular organisms, some cells specialize All rights reserved in providing sugars to other cells (e.g., intestinal and liver cells in animals, ∗ These authors contributed equally to this review. photosynthetic cells in plants), whereas others depend completely on an ex- Annu. Rev. Biochem. 2015.84:865-894. Downloaded from www.annualreviews.org ternal supply (e.g., brain cells, roots and seeds). This cellular exchange of Access provided by b-on: Universidade de Lisboa (UL) on 09/05/16. For personal use only. sugars requires transport proteins to mediate uptake or release from cells or subcellular compartments. Thus, not surprisingly, sugar transport is criti- cal for plants, animals, and humans. -

Supplemental Information
Supplemental information Dissection of the genomic structure of the miR-183/96/182 gene. Previously, we showed that the miR-183/96/182 cluster is an intergenic miRNA cluster, located in a ~60-kb interval between the genes encoding nuclear respiratory factor-1 (Nrf1) and ubiquitin-conjugating enzyme E2H (Ube2h) on mouse chr6qA3.3 (1). To start to uncover the genomic structure of the miR- 183/96/182 gene, we first studied genomic features around miR-183/96/182 in the UCSC genome browser (http://genome.UCSC.edu/), and identified two CpG islands 3.4-6.5 kb 5’ of pre-miR-183, the most 5’ miRNA of the cluster (Fig. 1A; Fig. S1 and Seq. S1). A cDNA clone, AK044220, located at 3.2-4.6 kb 5’ to pre-miR-183, encompasses the second CpG island (Fig. 1A; Fig. S1). We hypothesized that this cDNA clone was derived from 5’ exon(s) of the primary transcript of the miR-183/96/182 gene, as CpG islands are often associated with promoters (2). Supporting this hypothesis, multiple expressed sequences detected by gene-trap clones, including clone D016D06 (3, 4), were co-localized with the cDNA clone AK044220 (Fig. 1A; Fig. S1). Clone D016D06, deposited by the German GeneTrap Consortium (GGTC) (http://tikus.gsf.de) (3, 4), was derived from insertion of a retroviral construct, rFlpROSAβgeo in 129S2 ES cells (Fig. 1A and C). The rFlpROSAβgeo construct carries a promoterless reporter gene, the β−geo cassette - an in-frame fusion of the β-galactosidase and neomycin resistance (Neor) gene (5), with a splicing acceptor (SA) immediately upstream, and a polyA signal downstream of the β−geo cassette (Fig. -

Loss of the Tectorial Membrane Protein CEACAM16 Enhances Spontaneous, Stimulus-Frequency, and Transiently Evoked Otoacoustic Emissions
The Journal of Neuroscience, July 30, 2014 • 34(31):10325–10338 • 10325 Cellular/Molecular Loss of the Tectorial Membrane Protein CEACAM16 Enhances Spontaneous, Stimulus-Frequency, and Transiently Evoked Otoacoustic Emissions Mary Ann Cheatham,1 Richard J. Goodyear,3 Kazuaki Homma,4 P. Kevin Legan,3 Julia Korchagina,3 Souvik Naskar,3 Jonathan H. Siegel,1 X Peter Dallos,1,2 Jing Zheng,4 and Guy P. Richardson3 1Roxelyn and Richard Pepper Department of Communication Sciences and Disorders, The Knowles Hearing Center, and 2Department of Neurobiology, Northwestern University, Evanston, Illinois 60208, 3Sussex Neuroscience, School of Life Sciences, University of Sussex, Falmer, Brighton, BN1 9QG, United Kingdom, and 4Department of Otolaryngology-Head and Neck Surgery, The Knowles Hearing Center, Feinberg School of Medicine, Northwestern University, Chicago, Illinois 60611 ␣-Tectorin (TECTA), -tectorin (TECTB), and carcinoembryonic antigen-related cell adhesion molecule 16 (CEACAM) are secreted glycoproteins that are present in the tectorial membrane (TM), an extracellular structure overlying the hearing organ of the inner ear, the organ of Corti. Previous studies have shown that TECTA and TECTB are both required for formation of the striated-sheet matrix within which collagen fibrils of the TM are imbedded and that CEACAM16 interacts with TECTA. To learn more about the structural and functional significance of CEACAM16, we created a Ceacam16-null mutant mouse. In the absence of CEACAM16, TECTB levels are reduced, a clearly defined striated-sheet matrix does not develop, and Hensen’s stripe, a prominent feature in the basal two-thirds of the TM in WT mice, is absent. CEACAM16 is also shown to interact with TECTB, indicating that it may stabilize interactions between TECTA and TECTB. -

Role of Amylase in Ovarian Cancer Mai Mohamed University of South Florida, [email protected]
University of South Florida Scholar Commons Graduate Theses and Dissertations Graduate School July 2017 Role of Amylase in Ovarian Cancer Mai Mohamed University of South Florida, [email protected] Follow this and additional works at: http://scholarcommons.usf.edu/etd Part of the Pathology Commons Scholar Commons Citation Mohamed, Mai, "Role of Amylase in Ovarian Cancer" (2017). Graduate Theses and Dissertations. http://scholarcommons.usf.edu/etd/6907 This Dissertation is brought to you for free and open access by the Graduate School at Scholar Commons. It has been accepted for inclusion in Graduate Theses and Dissertations by an authorized administrator of Scholar Commons. For more information, please contact [email protected]. Role of Amylase in Ovarian Cancer by Mai Mohamed A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy Department of Pathology and Cell Biology Morsani College of Medicine University of South Florida Major Professor: Patricia Kruk, Ph.D. Paula C. Bickford, Ph.D. Meera Nanjundan, Ph.D. Marzenna Wiranowska, Ph.D. Lauri Wright, Ph.D. Date of Approval: June 29, 2017 Keywords: ovarian cancer, amylase, computational analyses, glycocalyx, cellular invasion Copyright © 2017, Mai Mohamed Dedication This dissertation is dedicated to my parents, Ahmed and Fatma, who have always stressed the importance of education, and, throughout my education, have been my strongest source of encouragement and support. They always believed in me and I am eternally grateful to them. I would also like to thank my brothers, Mohamed and Hussien, and my sister, Mariam. I would also like to thank my husband, Ahmed.