Marine Ecology Progress Series 546:239
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
HELCOM Red List
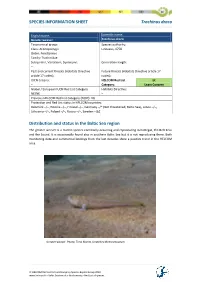
SPECIES INFORMATION SHEET Trachinus draco English name: Scientific name: Greater weever Trachinus draco Taxonomical group: Species authority: Class: Actinopterygii Linnaeus, 1758 Order: Perciformes Family: Trachinidae Subspecies, Variations, Synonyms: Generation length: – Past and current threats (Habitats Directive Future threats (Habitats Directive article 17 article 17 codes): – codes): – IUCN Criteria: HELCOM Red List LC – Category: Least Concern Global / European IUCN Red List Category Habitats Directive: NE/NE – Previous HELCOM Red List Category (2007): VU Protection and Red List status in HELCOM countries: Denmark –/–, Estonia –/–, Finland –/–, Germany –/* (Not threatened, Baltic Sea), Latvia –/–, Lithuania –/–, Poland –/–, Russia –/–, Sweden –/LC Distribution and status in the Baltic Sea region The greater weever is a marine species commonly occurring and reproducing in Kattegat, the Belt Seas and the Sound. It is occasionally found also in southern Baltic Sea but it is not reproducing there. Both monitoring data and commercial landings from the last decades show a positive trend in the HELCOM area. Greater weaver. Photo: Timo Moritz, Deutches Meeresmuseum. © HELCOM Red List Fish and Lamprey Species Expert Group 2013 www.helcom.fi > Baltic Sea trends > Biodiversity > Red List of species SPECIES INFORMATION SHEET Trachinus draco Fig.1 Catch per unit effort (number per trawling hour) of greater weever in international bottom trawl surveys in Kattegat (IBTS) during third quarter of the year (Linear fit and correlation coefficient -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

Atlas of North Sea Fishes
ICES COOPERATIVE RESEARCH REPORT RAPPORT DES RECHERCHES COLLECTIVES NO. 194 Atlas of North Sea Fishes Based on bottom-trawl survey data for the years 1985—1987 Ruud J. Knijn1, Trevor W. Boon2, Henk J. L. Heessen1, and John R. G. Hislop3 'Netherlands Institute for Fisheries Research, Haringkade 1, PO Box 6 8 , 1970 AB Umuiden, The Netherlands 2MAFF, Fisheries Laboratory, Lowestoft, Suffolk NR33 OHT, England 3Marine Laboratory, PO Box 101, Victoria Road, Aberdeen AB9 8 DB, Scotland Fish illustrations by Peter Stebbing International Council for the Exploration of the Sea Conseil International pour l’Exploration de la Mer Palægade 2—4, DK-1261 Copenhagen K, Denmark September 1993 Copyright ® 1993 All rights reserved No part of this book may be reproduced in any form by photostat or microfilm or stored in a storage system or retrieval system or by any other means without written permission from the authors and the International Council for the Exploration of the Sea Illustrations ® 1993 Peter Stebbing Published with financial support from the Directorate-General for Fisheries, AIR Programme, of the Commission of the European Communities ICES Cooperative Research Report No. 194 Atlas of North Sea Fishes ISSN 1017-6195 Printed in Denmark Contents 1. Introduction............................................................................................................... 1 2. Recruit surveys.................................................................................. 3 2.1 General purpose of the surveys..................................................................... -

Intrinsic Vulnerability in the Global Fish Catch
The following appendix accompanies the article Intrinsic vulnerability in the global fish catch William W. L. Cheung1,*, Reg Watson1, Telmo Morato1,2, Tony J. Pitcher1, Daniel Pauly1 1Fisheries Centre, The University of British Columbia, Aquatic Ecosystems Research Laboratory (AERL), 2202 Main Mall, Vancouver, British Columbia V6T 1Z4, Canada 2Departamento de Oceanografia e Pescas, Universidade dos Açores, 9901-862 Horta, Portugal *Email: [email protected] Marine Ecology Progress Series 333:1–12 (2007) Appendix 1. Intrinsic vulnerability index of fish taxa represented in the global catch, based on the Sea Around Us database (www.seaaroundus.org) Taxonomic Intrinsic level Taxon Common name vulnerability Family Pristidae Sawfishes 88 Squatinidae Angel sharks 80 Anarhichadidae Wolffishes 78 Carcharhinidae Requiem sharks 77 Sphyrnidae Hammerhead, bonnethead, scoophead shark 77 Macrouridae Grenadiers or rattails 75 Rajidae Skates 72 Alepocephalidae Slickheads 71 Lophiidae Goosefishes 70 Torpedinidae Electric rays 68 Belonidae Needlefishes 67 Emmelichthyidae Rovers 66 Nototheniidae Cod icefishes 65 Ophidiidae Cusk-eels 65 Trachichthyidae Slimeheads 64 Channichthyidae Crocodile icefishes 63 Myliobatidae Eagle and manta rays 63 Squalidae Dogfish sharks 62 Congridae Conger and garden eels 60 Serranidae Sea basses: groupers and fairy basslets 60 Exocoetidae Flyingfishes 59 Malacanthidae Tilefishes 58 Scorpaenidae Scorpionfishes or rockfishes 58 Polynemidae Threadfins 56 Triakidae Houndsharks 56 Istiophoridae Billfishes 55 Petromyzontidae -

Length-Weight Relationships of Marine Fish Collected from Around the British Isles
Science Series Technical Report no. 150 Length-weight relationships of marine fish collected from around the British Isles J. F. Silva, J. R. Ellis and R. A. Ayers Science Series Technical Report no. 150 Length-weight relationships of marine fish collected from around the British Isles J. F. Silva, J. R. Ellis and R. A. Ayers This report should be cited as: Silva J. F., Ellis J. R. and Ayers R. A. 2013. Length-weight relationships of marine fish collected from around the British Isles. Sci. Ser. Tech. Rep., Cefas Lowestoft, 150: 109 pp. Additional copies can be obtained from Cefas by e-mailing a request to [email protected] or downloading from the Cefas website www.cefas.defra.gov.uk. © Crown copyright, 2013 This publication (excluding the logos) may be re-used free of charge in any format or medium for research for non-commercial purposes, private study or for internal circulation within an organisation. This is subject to it being re-used accurately and not used in a misleading context. The material must be acknowledged as Crown copyright and the title of the publication specified. This publication is also available at www.cefas.defra.gov.uk For any other use of this material please apply for a Click-Use Licence for core material at www.hmso.gov.uk/copyright/licences/ core/core_licence.htm, or by writing to: HMSO’s Licensing Division St Clements House 2-16 Colegate Norwich NR3 1BQ Fax: 01603 723000 E-mail: [email protected] 3 Contents Contents 1. Introduction 5 2. -

ASFIS ISSCAAP Fish List February 2007 Sorted on Scientific Name
ASFIS ISSCAAP Fish List Sorted on Scientific Name February 2007 Scientific name English Name French name Spanish Name Code Abalistes stellaris (Bloch & Schneider 1801) Starry triggerfish AJS Abbottina rivularis (Basilewsky 1855) Chinese false gudgeon ABB Ablabys binotatus (Peters 1855) Redskinfish ABW Ablennes hians (Valenciennes 1846) Flat needlefish Orphie plate Agujón sable BAF Aborichthys elongatus Hora 1921 ABE Abralia andamanika Goodrich 1898 BLK Abralia veranyi (Rüppell 1844) Verany's enope squid Encornet de Verany Enoploluria de Verany BLJ Abraliopsis pfefferi (Verany 1837) Pfeffer's enope squid Encornet de Pfeffer Enoploluria de Pfeffer BJF Abramis brama (Linnaeus 1758) Freshwater bream Brème d'eau douce Brema común FBM Abramis spp Freshwater breams nei Brèmes d'eau douce nca Bremas nep FBR Abramites eques (Steindachner 1878) ABQ Abudefduf luridus (Cuvier 1830) Canary damsel AUU Abudefduf saxatilis (Linnaeus 1758) Sergeant-major ABU Abyssobrotula galatheae Nielsen 1977 OAG Abyssocottus elochini Taliev 1955 AEZ Abythites lepidogenys (Smith & Radcliffe 1913) AHD Acanella spp Branched bamboo coral KQL Acanthacaris caeca (A. Milne Edwards 1881) Atlantic deep-sea lobster Langoustine arganelle Cigala de fondo NTK Acanthacaris tenuimana Bate 1888 Prickly deep-sea lobster Langoustine spinuleuse Cigala raspa NHI Acanthalburnus microlepis (De Filippi 1861) Blackbrow bleak AHL Acanthaphritis barbata (Okamura & Kishida 1963) NHT Acantharchus pomotis (Baird 1855) Mud sunfish AKP Acanthaxius caespitosa (Squires 1979) Deepwater mud lobster Langouste -

Agnatha, Chondrichthyes and Osteichthyes
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/311861377 Agnatha, Chondrichthyes and Osteichthyes Chapter · November 2016 CITATIONS READS 0 1,531 2 authors: Antonis Petrou Charitos Zapitis AP Marine Env.Consultancy Ltd & Enalia Physis Environmental Research Centre University of Derby 29 PUBLICATIONS 131 CITATIONS 1 PUBLICATION 0 CITATIONS SEE PROFILE SEE PROFILE Some of the authors of this publication are also working on these related projects: Lionfish in the Mediterranean View project PCY1 - Investigating the effects of recreational diving on the macroalgal communities of the 'Zenobia' shipwreck (Cyprus) View project All content following this page was uploaded by Charitos Zapitis on 24 December 2016. The user has requested enhancement of the downloaded file. Chapter 26 - Agnatha CHAPTER 26: AGNATHA, CHONDRICHTHYES AND OSTEICHTHYES FISHES Antonis Petrou and Charitos Zapitis 1. INTRODUCTION The classification of fishes is not straightforward since they do not form a natural scientific grouping like the other vertebrate classes, i.e. the amphibians, reptiles, birds and mammals. Indeed, fishes can be considered by exclusion to be vertebrates that are not tetrapods (see Chapter 25: Introduction to Vertebrates). They are aquatic, gill- bearing, ectothermic ('cold-blooded') animals with a distinguishable head and, when present, digit-less limbs. Traditionally, fish have been arranged into three groups: ➵ Agnatha 1, the jawless fish (Myxini [hagfishes] and Hyperoartia [lampreys]); ➵ Chondrichthyes, the cartilaginous fish (sharks, skates and rays); ➵ Osteichthyes, the bony fish (Actinopterygii [ray-finned fishes] and Sarcopterygii [lobe-finned fishes]). This classification is adequate for general purposes, although Agnatha is paraphyletic and includes several groups of extinct jawless fishes. -

Bluefin Tuna Larval Survey 2008 Oceana-Marviva Mediterranean Project · 2008 Oceana-Marviva
Plaza de España - Leganitos, 47 28013 Madrid (Spain) Tel.: + 34 911 440 880 Fax: + 34 911 440 890 [email protected] www.oceana.org Rue Montoyer, 39 1000 Brussels (Belgium) Tel.: + 32 (0) 2 513 22 42 Fax: + 32 (0) 2 513 22 46 [email protected] 1350 Connecticut Ave., NW, 5th Floor Washington D.C., 20036 USA Tel.: + 1 (202) 833 3900 Fax: + 1 (202) 833 2070 [email protected] 175 South Franklin Street - Suite 418 Juneau, Alaska 99801 (USA) Tel.: + 1 (907) 586 40 50 Fax: + 1(907) 586 49 44 [email protected] Avenida General Bustamante, 24, Departamento 2C 750-0776 Providencia, Santiago (Chile) Tel.: + 56 2 795 7140 Fax: + 56 2 795 7146 [email protected] Bluefin Tuna Larval Survey 2008 Oceana-MarViva Mediterranean Project 2008 Oceana-MarViva · 2008 Oceana-MarViva Mediterranean Project Bluefin Tuna Larval Survey Bluefin The research included in this report and its publication were carried out by Oceana with the support of Fundación MarViva. Project Director • Xavier Pastor Authors • Ricardo Aguilar, Patricia Lastra Editor • Marta Madina Editorial Assistants • Rebecca Greenberg, Aitor Lascurain, Ángeles Sáez, Natividad Sánchez Cover • Larvae sample under microscope lens. © OCEANA/ Eduardo de Ana Design and Layout • NEO Estudio Gráfico, S.L. Printer • Imprenta Roal, S.L. Photo Montage • Pentados, S.A. Acknowledgements • Oceana would like to thank the Spanish Oceanographic Institute (Instituto Español de Oceanografía, IEO), and especially Francisco Alemany, IEO investigator, for his scientific advice. Oceana would also like to show its appreciation for the support received from the Marviva Med crew, in particular from Carlos Pérez and César Fuertes. -

Trachinus Draco 5A
6 Bony Fishes Perciformes: Trachinoidei: Trachinidae TRACHINIDAE Weeverfishes by W.L. Smith, The University of Kansas, Lawrence, KS, USA (after Roux, 1981, 1990) iagnostic characters: Elongate, rather compressed, moderate-sized (to 45 cm total length) perciform Dfishes. Eyes small (eye diameter contained 4 to 5 times in head length), approaching or protruding slightly above dorsal profile of head; head small with a short snout; mouth large, strongly oblique, extending to posterior margin of eye or just beyond when mouth is closed; villiform teeth in jaws and on palate; vomerine teeth absent or present. Dorsal fins separate, first fin short with 5 to 8 spines; second fin long with 0 spines and 21 to 32 segmented soft rays; anal fin long with 1 or 2 spines and 24 to 34 segmented soft rays. Pectoral fins with 15 rays. Pelvic fins in advance of and smaller than pectoral fins. Large, venomous spine on gill cover and venomous dorsal spines. Vertebrae 34 to 43. Body covered with small, cycloid or ctenoid scales in oblique series; lateral line straight, occasionally ventrally displaced on caudal peduncle. Gas bladder absent. Colour: variable across species, ranging from off-white to greenish yellow to dark brown; most species have spots, oblique lines, or reticulations. venomous spine on long second dorsal fin gill cover large mouth, strongly oblique Habitat, biology, and fisheries: Littoral or benthic fishes inhabiting sandy or muddy bottoms on the continental shelf (occasionally collected deeper between 150 and 200 m). They are frequently encountered buried in the sediment with their eyes and venomous dorsal spines exposed. -

Four Important Poisonous Fishes in the Black Sea Coasts: Length-Weight Relationships (Lwrs) and Some Biological Features
Journal of New Results in Science (JNRS) Volume: 9 Issue: 3 ISSN: 1304-7981 http://dergipark.gov.tr/en/pub/jnrs Year: 2020 Research Article Pages: 16-24 Open Access Received: 22.09.2020 Accepted: 03.12.2020 Four Important Poisonous Fishes in the Black Sea Coasts: Length-Weight Relationships (LWRs) and Some Biological Features Süleyman Özdemira,*, Ferhat Büyükdevecib, Uğur Özsandıkçıa, Yakup Erdema a Department of Fishing Technology, Faculty of Fisheries, Sinop University, Sinop, Turkey b District Directorate of Agriculture and Forestry of Sarıçam, Republic of Turkey Ministry of Agriculture and Forestry, Adana, Turkey *Corresponding author, [email protected] ABSTRACT: In this study it was demonstrated that length-weight relationships (LWRs) and some biological characteristic of four venomous fishes, common stingray (Dasyatis pastinaca), greater weever (Trachinus draco), scorpion fish (Scorpaena porcus) and stargazer (Uranoscopus scaber) from Black Sea coasts. The present study was carried out between 01 January 2015 and 30 September 2016 in Sinop peninsula coasts of the southern Black Sea. The fish samples were captured trammel nets (32 mm, 36 mm 40 mm and 48 mm mesh size) in 0-15-30-45 meters depth ranges. The average length of common stingray, greater weever, scorpion fish and stargazer were established 48.2±4.98 cm 17.88±0.25 cm, 11.49±0.07 cm and 13.25±0.47 cm respectively. LWR of the venomous fishes were determined W=0.0028L3.2187 (n=12, R2=0.97), W=0.0287L2.5776 (n= 181, R2=0.93), W=0.0245L2.9050 (n=808, R2=0.98) and W=0.0181L2.9398 (n=779, R2=0.98), respectively. -
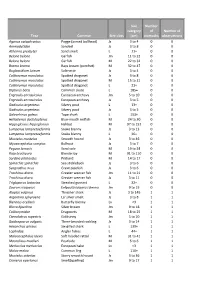
Taxa Common Size Class Size Category (Cm) Number of Stomachs
Size Number category of Number of Taxa Common Size class (cm) stomachs observations Agonus cataphractus Pogge (armed bullhead) Js 3 to 4 0 0 Ammodytidae Sandeel Js 3 to 8 0 0 Atherina presbyter Sand smelt L 11+ 0 0 Belone belone Garfish Jm 11 to 22 0 0 Belone belone Garfish M 22 to 32 0 0 Brama brama Rays bream (pomfret) M 32 to 47 0 0 Buglossidium luteum Solenette Js 3 to 4 0 0 Callionymus maculatus Spotted dragonet Js 3 to 8 0 0 Callionymus maculatus Spotted dragonet M 15 to 21 0 0 Callionymus maculatus Spotted dragonet L 21+ 0 0 Dipturus batis Common skate L 185+ 0 0 Engraulis encrasicolus European anchovy Jm 5 to 10 0 0 Engraulis encrasicolus European anchovy Js 3 to 5 0 0 Gadiculus argenteus Silvery pout L 13+ 0 0 Gadiculus argenteus Silvery pout Js 3 to 5 0 0 Galeorhinus galeus Tope shark L 153+ 0 0 Helicolenus dactylopterus Blue-mouth redfish M 24 to 30 0 0 Hippoglossus hippoglossus Halibut M 97 to 151 0 0 Lumpenus lampretaeformis Snake blenny Js 3 to 13 0 0 Lumpenus lampretaeformis Snake blenny L 36+ 0 0 Mustelus mustelus Smooth hound Js 3 to 46 0 0 Myoxocephalus scorpius Bullrout Js 3 to 7 0 0 Pegusa lascaris Sand sole M 19 to 24 0 0 Raja brachyura Blonde ray M 91 to 110 0 0 Sardina pilchardus Pilchard M 14 to 17 0 0 Spinachia spinachia Sea stickleback Js 3 to 6 0 0 Syngnathus acus Great pipefish Js 3 to 6 0 0 Trachinus draco Greater weever fish Jm 11 to 21 0 0 Trachinus draco Greater weever fish Js 3 to 11 0 0 Trigloporus lastoviza Streaked gurnard L 32+ 0 0 Zoarces viviparus Eelpout/viviparus blenny Jm 9 to 19 0 0 Alopias vulpinus -

Wkcofibyc 2021
WORKSHOP ON FISH OF CONSERVATION AND BYCATCH RELEVANCE (WKCOFIBYC; outputs from 2020 meeting) VOLUME 3 | ISSUE 57 ICES SCIENTIFIC REPORTS RAPPORTS SCIENTIFIQUES DU CIEM ICES INTERNATIONAL COUNCIL FOR THE EXPLORATION OF THE SEA CIEM CONSEIL INTERNATIONAL POUR L’EXPLORATION DE LA MER International Council for the Exploration of the Sea Conseil International pour l’Exploration de la Mer H.C. Andersens Boulevard 44-46 DK-1553 Copenhagen V Denmark Telephone (+45) 33 38 67 00 Telefax (+45) 33 93 42 15 www.ices.dk [email protected] ISSN number: 2618-1371 This document has been produced under the auspices of an ICES Expert Group or Committee. The contents therein do not necessarily represent the view of the Council. © 2021 International Council for the Exploration of the Sea. This work is licensed under the Creative Commons Attribution 4.0 International License (CC BY 4.0). For citation of datasets or conditions for use of data to be included in other databases, please refer to ICES data policy. ICES Scientific Reports Volume 3 | Issue 57 WORKSHOP ON FISH OF CONSERVATION AND BYCATCH RELEVANCE (WKCOFIBYC) Recommended format for purpose of citation: ICES. 2021. Workshop on Fish of Conservation and Bycatch Relevance (WKCOFIBYC). ICES Scientific Reports. 3:57. 125 pp. https://doi.org/10.17895/ices.pub.8194 Editors Maurice Clarke Authors Sara Bonanomi • Archontia Chatzispyrou • Maurice Clarke • Bram Couperus • Jim Ellis • Ruth Fernández • Ailbhe Kavanagh • Allen Kingston • Vasiliki Kousteni • Evgenia Lefkaditou • Henn Ojaveer Wolfgang Nikolaus Probst •