Characterisation of Marine Toxins with Emphasis on the Neglected Status of Marine Toxinology
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
HELCOM Red List
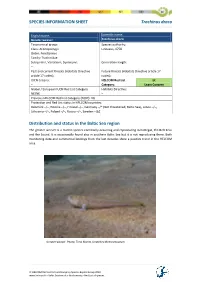
SPECIES INFORMATION SHEET Trachinus draco English name: Scientific name: Greater weever Trachinus draco Taxonomical group: Species authority: Class: Actinopterygii Linnaeus, 1758 Order: Perciformes Family: Trachinidae Subspecies, Variations, Synonyms: Generation length: – Past and current threats (Habitats Directive Future threats (Habitats Directive article 17 article 17 codes): – codes): – IUCN Criteria: HELCOM Red List LC – Category: Least Concern Global / European IUCN Red List Category Habitats Directive: NE/NE – Previous HELCOM Red List Category (2007): VU Protection and Red List status in HELCOM countries: Denmark –/–, Estonia –/–, Finland –/–, Germany –/* (Not threatened, Baltic Sea), Latvia –/–, Lithuania –/–, Poland –/–, Russia –/–, Sweden –/LC Distribution and status in the Baltic Sea region The greater weever is a marine species commonly occurring and reproducing in Kattegat, the Belt Seas and the Sound. It is occasionally found also in southern Baltic Sea but it is not reproducing there. Both monitoring data and commercial landings from the last decades show a positive trend in the HELCOM area. Greater weaver. Photo: Timo Moritz, Deutches Meeresmuseum. © HELCOM Red List Fish and Lamprey Species Expert Group 2013 www.helcom.fi > Baltic Sea trends > Biodiversity > Red List of species SPECIES INFORMATION SHEET Trachinus draco Fig.1 Catch per unit effort (number per trawling hour) of greater weever in international bottom trawl surveys in Kattegat (IBTS) during third quarter of the year (Linear fit and correlation coefficient -

Sharkcam Fishes
SharkCam Fishes A Guide to Nekton at Frying Pan Tower By Erin J. Burge, Christopher E. O’Brien, and jon-newbie 1 Table of Contents Identification Images Species Profiles Additional Info Index Trevor Mendelow, designer of SharkCam, on August 31, 2014, the day of the original SharkCam installation. SharkCam Fishes. A Guide to Nekton at Frying Pan Tower. 5th edition by Erin J. Burge, Christopher E. O’Brien, and jon-newbie is licensed under the Creative Commons Attribution-Noncommercial 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc/4.0/. For questions related to this guide or its usage contact Erin Burge. The suggested citation for this guide is: Burge EJ, CE O’Brien and jon-newbie. 2020. SharkCam Fishes. A Guide to Nekton at Frying Pan Tower. 5th edition. Los Angeles: Explore.org Ocean Frontiers. 201 pp. Available online http://explore.org/live-cams/player/shark-cam. Guide version 5.0. 24 February 2020. 2 Table of Contents Identification Images Species Profiles Additional Info Index TABLE OF CONTENTS SILVERY FISHES (23) ........................... 47 African Pompano ......................................... 48 FOREWORD AND INTRODUCTION .............. 6 Crevalle Jack ................................................. 49 IDENTIFICATION IMAGES ...................... 10 Permit .......................................................... 50 Sharks and Rays ........................................ 10 Almaco Jack ................................................. 51 Illustrations of SharkCam -

Revisión Taxonómica De La Ictiología Marina De Galicia: Clase Actinopteri (Orden Trachiniformes Al Orden Tetraodontiformes)
Nova Acta Científica Compostelana (Bioloxía), 28: 77-104 (2021) - ISSN 2340-0021 ARTÍCULO DE INVESTIGACIÓN Revisión taxonómica de la ictiología marina de Galicia: Clase Actinopteri (Orden Trachiniformes al Orden Tetraodontiformes) Taxonomic review of Galician marine ichthyology: Classe Actinopteri (Order Trachiniformes to Order Tetraodontiformes) *RAFAEL BAÑÓN, TOÑO MAÑO Grupo de Estudos do Medio Mariño (GEMM), Puerto Deportivo s/n 15960 Ribeira, A Coruña, España. *[email protected]; [email protected] (Recibido 29/11/2020; Aceptado 26/03/2021) Resumen En este trabajo se realiza una revisión taxonómica de los peces óseos de Galicia (Clase Actinopteri) del Orden Trachiniformes al Orden Tetraodontiformes, a través de los distintos tratados y publicaciones ictio- lógicas publicadas a lo largo de la historia. Se listan un total de 188 especies, de las cuales 5 se consideran dudosas, al no estar su presencia suficientemente demostrada. Una revisión de la bibliografía y nomenclatura científica nos ha permitido citar nuevas especies para Galicia y reasignar antiguas denominaciones a nuevas especies, subsanando errores de identificación de otros autores. El orden Perciformes, con 145 especies, es el más numeroso de los peces de Galicia. A este orden pertenecen especies de alto interés comercial como el jurel Trachurus trachurus y la caballa Scomber scombrus. El listado incluye también los primeros registros para Galicia de especies de carácter tropical desplazadas hacia el norte debido al cambio climático a lo largo de estas últimas décadas. Algunas de estas especies son el jurelo azul Caranx crysos, el pez globo Lagocephalus laevigatus y el mero tropical Epinephelus aeneus. Palabras clave: Peces óseos, nomenclatura, ictiología, tropicalización. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

New Zealand Fishes a Field Guide to Common Species Caught by Bottom, Midwater, and Surface Fishing Cover Photos: Top – Kingfish (Seriola Lalandi), Malcolm Francis
New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing Cover photos: Top – Kingfish (Seriola lalandi), Malcolm Francis. Top left – Snapper (Chrysophrys auratus), Malcolm Francis. Centre – Catch of hoki (Macruronus novaezelandiae), Neil Bagley (NIWA). Bottom left – Jack mackerel (Trachurus sp.), Malcolm Francis. Bottom – Orange roughy (Hoplostethus atlanticus), NIWA. New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing New Zealand Aquatic Environment and Biodiversity Report No: 208 Prepared for Fisheries New Zealand by P. J. McMillan M. P. Francis G. D. James L. J. Paul P. Marriott E. J. Mackay B. A. Wood D. W. Stevens L. H. Griggs S. J. Baird C. D. Roberts‡ A. L. Stewart‡ C. D. Struthers‡ J. E. Robbins NIWA, Private Bag 14901, Wellington 6241 ‡ Museum of New Zealand Te Papa Tongarewa, PO Box 467, Wellington, 6011Wellington ISSN 1176-9440 (print) ISSN 1179-6480 (online) ISBN 978-1-98-859425-5 (print) ISBN 978-1-98-859426-2 (online) 2019 Disclaimer While every effort was made to ensure the information in this publication is accurate, Fisheries New Zealand does not accept any responsibility or liability for error of fact, omission, interpretation or opinion that may be present, nor for the consequences of any decisions based on this information. Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries website at http://www.mpi.govt.nz/news-and-resources/publications/ A higher resolution (larger) PDF of this guide is also available by application to: [email protected] Citation: McMillan, P.J.; Francis, M.P.; James, G.D.; Paul, L.J.; Marriott, P.; Mackay, E.; Wood, B.A.; Stevens, D.W.; Griggs, L.H.; Baird, S.J.; Roberts, C.D.; Stewart, A.L.; Struthers, C.D.; Robbins, J.E. -

A Check-List of Polychaete Species from the Black
J. Black Sea/Mediterranean Environment Vol. 18, No. 1: 76-82 (2012) SHORT COMMUNICATION First sighting of the Red Sea originated stonefish (Synanceia verrucosa) from Turkey Murat Bilecenoğlu* Department of Biology, Faculty of Arts and Sciences, Adnan Menderes University, 09010, Aydın, TURKEY *Corresponding author: [email protected] Abstract A single specimen of stonefish (Synanceia verrucosa Bloch and Schneider, 1801) was recently captured off Yumurtalık (Iskenderun Bay), representing its first occurrence on the coast of Turkey and second record in the entire Mediterranean Sea. This species is famous with its highly toxic stings and possess a potential risk for human health, if it finds the opportunity to establish a successfully breeding population in the region. Key words: Synanceia verrucosa, Synanceiidae, alien species, Mediterranean Sea, Turkey Introduction The number of alien species in the Mediterranean has currently reached to the psychological threshold of 1000 species (Zenetos et al. 2010), advancing the rank of this semi-enclosed sea as one of the most invaded ecosystems on Earth. Despite of the widespread “native good, alien bad” philosophy (Goodenough 2010), ecological and sociological impacts of alien species have proven to share both sides of this approach (see Cinar et al. 2011). This paper deals with a newly introduced venomous fish species on the northeastern Levantine coast of Turkey, which doubtless takes part in the negative wing among alien biota. Species diagnosis On 18 November 2011, a single specimen of stonefish (Synanceia verrucosa Bloch and Schneider, 1801) was captured along the Yumurtalık coast (Adana, Iskenderun Bay) by an artisanal fisherman, presumably using a bottom long- line. -

Venom Week 2012 4Th International Scientific Symposium on All Things Venomous
17th World Congress of the International Society on Toxinology Animal, Plant and Microbial Toxins & Venom Week 2012 4th International Scientific Symposium on All Things Venomous Honolulu, Hawaii, USA, July 8 – 13, 2012 1 Table of Contents Section Page Introduction 01 Scientific Organizing Committee 02 Local Organizing Committee / Sponsors / Co-Chairs 02 Welcome Messages 04 Governor’s Proclamation 08 Meeting Program 10 Sunday 13 Monday 15 Tuesday 20 Wednesday 26 Thursday 30 Friday 36 Poster Session I 41 Poster Session II 47 Supplemental program material 54 Additional Abstracts (#298 – #344) 61 International Society on Thrombosis & Haemostasis 99 2 Introduction Welcome to the 17th World Congress of the International Society on Toxinology (IST), held jointly with Venom Week 2012, 4th International Scientific Symposium on All Things Venomous, in Honolulu, Hawaii, USA, July 8 – 13, 2012. This is a supplement to the special issue of Toxicon. It contains the abstracts that were submitted too late for inclusion there, as well as a complete program agenda of the meeting, as well as other materials. At the time of this printing, we had 344 scientific abstracts scheduled for presentation and over 300 attendees from all over the planet. The World Congress of IST is held every three years, most recently in Recife, Brazil in March 2009. The IST World Congress is the primary international meeting bringing together scientists and physicians from around the world to discuss the most recent advances in the structure and function of natural toxins occurring in venomous animals, plants, or microorganisms, in medical, public health, and policy approaches to prevent or treat envenomations, and in the development of new toxin-derived drugs. -

Marine Fishes of the Azores: an Annotated Checklist and Bibliography
MARINE FISHES OF THE AZORES: AN ANNOTATED CHECKLIST AND BIBLIOGRAPHY. RICARDO SERRÃO SANTOS, FILIPE MORA PORTEIRO & JOÃO PEDRO BARREIROS SANTOS, RICARDO SERRÃO, FILIPE MORA PORTEIRO & JOÃO PEDRO BARREIROS 1997. Marine fishes of the Azores: An annotated checklist and bibliography. Arquipélago. Life and Marine Sciences Supplement 1: xxiii + 242pp. Ponta Delgada. ISSN 0873-4704. ISBN 972-9340-92-7. A list of the marine fishes of the Azores is presented. The list is based on a review of the literature combined with an examination of selected specimens available from collections of Azorean fishes deposited in museums, including the collection of fish at the Department of Oceanography and Fisheries of the University of the Azores (Horta). Personal information collected over several years is also incorporated. The geographic area considered is the Economic Exclusive Zone of the Azores. The list is organised in Classes, Orders and Families according to Nelson (1994). The scientific names are, for the most part, those used in Fishes of the North-eastern Atlantic and the Mediterranean (FNAM) (Whitehead et al. 1989), and they are organised in alphabetical order within the families. Clofnam numbers (see Hureau & Monod 1979) are included for reference. Information is given if the species is not cited for the Azores in FNAM. Whenever available, vernacular names are presented, both in Portuguese (Azorean names) and in English. Synonyms, misspellings and misidentifications found in the literature in reference to the occurrence of species in the Azores are also quoted. The 460 species listed, belong to 142 families; 12 species are cited for the first time for the Azores. -

Venom Evolution Widespread in Fishes: a Phylogenetic Road Map for the Bioprospecting of Piscine Venoms
Journal of Heredity 2006:97(3):206–217 ª The American Genetic Association. 2006. All rights reserved. doi:10.1093/jhered/esj034 For permissions, please email: [email protected]. Advance Access publication June 1, 2006 Venom Evolution Widespread in Fishes: A Phylogenetic Road Map for the Bioprospecting of Piscine Venoms WILLIAM LEO SMITH AND WARD C. WHEELER From the Department of Ecology, Evolution, and Environmental Biology, Columbia University, 1200 Amsterdam Avenue, New York, NY 10027 (Leo Smith); Division of Vertebrate Zoology (Ichthyology), American Museum of Natural History, Central Park West at 79th Street, New York, NY 10024-5192 (Leo Smith); and Division of Invertebrate Zoology, American Museum of Natural History, Central Park West at 79th Street, New York, NY 10024-5192 (Wheeler). Address correspondence to W. L. Smith at the address above, or e-mail: [email protected]. Abstract Knowledge of evolutionary relationships or phylogeny allows for effective predictions about the unstudied characteristics of species. These include the presence and biological activity of an organism’s venoms. To date, most venom bioprospecting has focused on snakes, resulting in six stroke and cancer treatment drugs that are nearing U.S. Food and Drug Administration review. Fishes, however, with thousands of venoms, represent an untapped resource of natural products. The first step in- volved in the efficient bioprospecting of these compounds is a phylogeny of venomous fishes. Here, we show the results of such an analysis and provide the first explicit suborder-level phylogeny for spiny-rayed fishes. The results, based on ;1.1 million aligned base pairs, suggest that, in contrast to previous estimates of 200 venomous fishes, .1,200 fishes in 12 clades should be presumed venomous. -

Atlas of North Sea Fishes
ICES COOPERATIVE RESEARCH REPORT RAPPORT DES RECHERCHES COLLECTIVES NO. 194 Atlas of North Sea Fishes Based on bottom-trawl survey data for the years 1985—1987 Ruud J. Knijn1, Trevor W. Boon2, Henk J. L. Heessen1, and John R. G. Hislop3 'Netherlands Institute for Fisheries Research, Haringkade 1, PO Box 6 8 , 1970 AB Umuiden, The Netherlands 2MAFF, Fisheries Laboratory, Lowestoft, Suffolk NR33 OHT, England 3Marine Laboratory, PO Box 101, Victoria Road, Aberdeen AB9 8 DB, Scotland Fish illustrations by Peter Stebbing International Council for the Exploration of the Sea Conseil International pour l’Exploration de la Mer Palægade 2—4, DK-1261 Copenhagen K, Denmark September 1993 Copyright ® 1993 All rights reserved No part of this book may be reproduced in any form by photostat or microfilm or stored in a storage system or retrieval system or by any other means without written permission from the authors and the International Council for the Exploration of the Sea Illustrations ® 1993 Peter Stebbing Published with financial support from the Directorate-General for Fisheries, AIR Programme, of the Commission of the European Communities ICES Cooperative Research Report No. 194 Atlas of North Sea Fishes ISSN 1017-6195 Printed in Denmark Contents 1. Introduction............................................................................................................... 1 2. Recruit surveys.................................................................................. 3 2.1 General purpose of the surveys..................................................................... -

APPENDIX 1 Classified List of Fishes Mentioned in the Text, with Scientific and Common Names
APPENDIX 1 Classified list of fishes mentioned in the text, with scientific and common names. ___________________________________________________________ Scientific names and classification are from Nelson (1994). Families are listed in the same order as in Nelson (1994), with species names following in alphabetical order. The common names of British fishes mostly follow Wheeler (1978). Common names of foreign fishes are taken from Froese & Pauly (2002). Species in square brackets are referred to in the text but are not found in British waters. Fishes restricted to fresh water are shown in bold type. Fishes ranging from fresh water through brackish water to the sea are underlined; this category includes diadromous fishes that regularly migrate between marine and freshwater environments, spawning either in the sea (catadromous fishes) or in fresh water (anadromous fishes). Not indicated are marine or freshwater fishes that occasionally venture into brackish water. Superclass Agnatha (jawless fishes) Class Myxini (hagfishes)1 Order Myxiniformes Family Myxinidae Myxine glutinosa, hagfish Class Cephalaspidomorphi (lampreys)1 Order Petromyzontiformes Family Petromyzontidae [Ichthyomyzon bdellium, Ohio lamprey] Lampetra fluviatilis, lampern, river lamprey Lampetra planeri, brook lamprey [Lampetra tridentata, Pacific lamprey] Lethenteron camtschaticum, Arctic lamprey] [Lethenteron zanandreai, Po brook lamprey] Petromyzon marinus, lamprey Superclass Gnathostomata (fishes with jaws) Grade Chondrichthiomorphi Class Chondrichthyes (cartilaginous -

Pterapogon Kauderni in Appendix II, in Accordance with Article II, Paragraph 2(A) of the Convention and Satisfying Criteria a and B in Annex 2A of Resolution Conf
Original language: English CoP17 Prop. XXX CONVENTION ON INTERNATIONAL TRADE IN ENDANGERED SPECIES OF WILD FAUNA AND FLORA ____________________ Seventeenth meeting of the Conference of the Parties Johannesburg (South Africa), 24 September – 5 October 2016 CONSIDERATION OF PROPOSALS FOR AMENDMENT OF APPENDICES I AND II A. Proposal Inclusion of Pterapogon kauderni in Appendix II, in accordance with Article II, paragraph 2(a) of the Convention and satisfying Criteria A and B in Annex 2a of Resolution Conf. 9.24 (Rev. CoP16). B. Proponent The European Union and its Member States* C. Supporting statement 1. Taxonomy 1.1 Class: Actinopterygii 1.2 Order: Perciformes 1.3 Family: Apogonidae 1.4 Genus, species or subspecies, including author and year: Pterapogon kauderni Koumans, 1933 1.5 Scientific synonyms: 1.6 Common names: English: Banggai Cardinalfish French: Poisson-cardinal de Banggai Spanish: Pez cardenal de Banggai 1.7 Code numbers: 2. Overview Pterapogon kauderni is a small marine fish endemic to the Banggai Archipelago off Central Sulawesi, eastern Indonesia (Allen and Steene, 2005; Vagelli and Erdmann, 2002). The species has an extremely restricted range of c. 5,500 km2 and occurs as isolated small populations in the shallows of 34 islands (Vagelli, 2011). The species has been subject to heavy collection pressure for the aquarium trade, with annual harvests reportedly having reached 900.000 fish/year in 2007 (Vagelli, 2008; 2011). The species’ biological characteristics make it vulnerable to overexploitation (low fecundity, extended parental care, and a lack of planktonic phase that precludes dispersal). A reported widespread decline in the abundance of * The geographical designations employed in this document do not imply the expression of any opinion whatsoever on the part of the CITES Secretariat (or the United Nations Environment Programme) concerning the legal status of any country, territory, or area, or concerning the delimitation of its frontiers or boundaries.