The Sequence and Function Relationship of Elastin: How
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A Database Integrating Genomic Indel Polymorphisms That Distinguish Mouse Strains Keiko Akagi1,2, Robert M
D600–D606 Nucleic Acids Research, 2010, Vol. 38, Database issue Published online 20 November 2009 doi:10.1093/nar/gkp1046 MouseIndelDB: a database integrating genomic indel polymorphisms that distinguish mouse strains Keiko Akagi1,2, Robert M. Stephens3, Jingfeng Li2,4, Evgenji Evdokimov5, Michael R. Kuehn5, Natalia Volfovsky3 and David E. Symer2,4,6,7,8,9,* 1Mouse Cancer Genetics Program, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, 2Department of Molecular Virology, Immunology and Medical Genetics, The Ohio State University Comprehensive Cancer Center, Columbus, OH 43210, 3Advanced Biomedical Computing Center, Information Systems Program, SAIC-Frederick, Inc., NCI-Frederick, Frederick, MD 21702, 4Basic Research Laboratory, 5Laboratory of Protein Dynamics and Signaling, 6Laboratory of Biochemistry and Molecular Biology, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, 7Human Cancer Genetics Program, 8Departments of Internal Medicine and 9Biomedical Informatics, The Ohio State University Comprehensive Cancer Center, Columbus, OH 43210, USA Received August 5, 2009; Revised October 23, 2009; Accepted October 27, 2009 ABSTRACT wide-ranging functional differences. This extensive phenotypic variation has helped to make the mouse a MouseIndelDB is an integrated database resource premier model organism, mimicking many aspects of containing thousands of previously unreported human diversity and diseases. Understanding the mouse genomic indel (insertion and deletion) genomic differences that -

Pathway in Protective Immunity Jeremy Manry, Guillaume Laval, Etienne Patin, Simona Fornarino, Christiane Bouchier, Magali Tichit, Luis Barreiro, Lluis Quintana-Murci
Evolutionary genetics evidence of an essential, non-redundant role of the IFN-γ pathway in protective immunity Jeremy Manry, Guillaume Laval, Etienne Patin, Simona Fornarino, Christiane Bouchier, Magali Tichit, Luis Barreiro, Lluis Quintana-Murci To cite this version: Jeremy Manry, Guillaume Laval, Etienne Patin, Simona Fornarino, Christiane Bouchier, et al.. Evo- lutionary genetics evidence of an essential, non-redundant role of the IFN-γ pathway in protective immunity. Human Mutation, Wiley, 2011, 32 (6), pp.633-42. 10.1002/humu.21484. hal-00627541 HAL Id: hal-00627541 https://hal.archives-ouvertes.fr/hal-00627541 Submitted on 29 Sep 2011 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Human Mutation Evolutionary genetics evidence of an essential, non- redundant role of the IFN-γ pathway in protective immunity For Peer Review Journal: Human Mutation Manuscript ID: humu-2010-0462.R1 Wiley - Manuscript type: Research Article Date Submitted by the 23-Jan-2011 Author: Complete List of Authors: Manry, Jeremy; Institut Pasteur Laval, Guillaume; Institut Pasteur Patin, Etienne; Institut Pasteur Fornarino, Simona; Institut Pasteur Bouchier, Christiane; Institut Pasteur Tichit, Magali; Institut Pasteur Barreiro, Luis; University of Chicago Quintana-Murci, Lluis; Institut Pasteur, EEMI IFNG, IFNGR1, IFNGR2, natural selection, polymorphism, Key Words: population genetics John Wiley & Sons, Inc. -
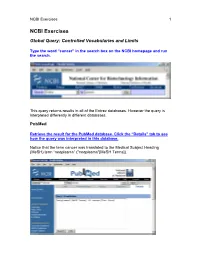
NCBI Exercises 1
NCBI Exercises 1 NCBI Exercises Global Query: Controlled Vocabularies and Limits Type the word “cancer” in the search box on the NCBI homepage and run the search. This query returns results in all of the Entrez databases. However the query is interpreted differently in different databases. PubMed Retrieve the result for the PubMed database. Click the “Details” tab to see how the query was interpreted in this database. Notice that the term cancer was translated to the Medical Subject Heading (MeSH) term “neoplasms” ("neoplasms"[MeSH Terms]). NCBI Exercises 2 MeSH is a controlled vocabulary that is used to index all articles in PubMed. In the details box, edit the query to remove the portion that searched for cancer as a text word and run the search. Notice that the number of articles retrieved has changed. These will be a more relevant set of results. You can force the PubMed engine to only search the MeSH vocabulary or specify any other indexed field through the “Limits” tab. Use the Web browser’s back button to return to the Global query page and retrieve the PubMed results again. Now click on the “Limits” tab. Select “MeSH terms” from the first drop-down menu, the one headed by “All Fields”. Now run the search with the limit in place and check the “Details” tab to verify that only the MeSH term translation was used. Nucleotide Use the Web browser’s back button to return to the Global query page. Retrieve the results for the nucleotide database. Click the “Details” tab to see how the query was interpreted for this molecular database. -

Mechanism of Resilin Elasticity
ARTICLE Received 24 Oct 2011 | Accepted 11 Jul 2012 | Published 14 Aug 2012 DOI: 10.1038/ncomms2004 Mechanism of resilin elasticity Guokui Qin1,*, Xiao Hu1,*, Peggy Cebe2 & David L. Kaplan1 Resilin is critical in the flight and jumping systems of insects as a polymeric rubber-like protein with outstanding elasticity. However, insight into the underlying molecular mechanisms responsible for resilin elasticity remains undefined. Here we report the structure and function of resilin from Drosophila CG15920. A reversible beta-turn transition was identified in the peptide encoded by exon III and for full-length resilin during energy input and release, features that correlate to the rapid deformation of resilin during functions in vivo. Micellar structures and nanoporous patterns formed after beta-turn structures were present via changes in either the thermal or the mechanical inputs. A model is proposed to explain the super elasticity and energy conversion mechanisms of resilin, providing important insight into structure–function relationships for this protein. Furthermore, this model offers a view of elastomeric proteins in general where beta-turn-related structures serve as fundamental units of the structure and elasticity. 1 Department of Biomedical Engineering, Tufts University, Medford, Massachusetts 02155, USA. 2 Department of Physics and Astronomy, Tufts University, Medford, Massachusetts 02155, USA. *These authors contributed equally to this work. Correspondence and requests for materials should be addressed to D.L.K. (email: [email protected]). -
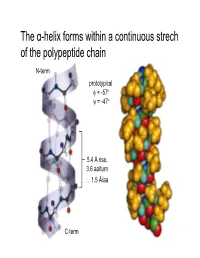
The Α-Helix Forms Within a Continuous Strech of the Polypeptide Chain
The α-helix forms within a continuous strech of the polypeptide chain N-term prototypical φ = -57 ° ψ = -47 ° 5.4 Å rise, 3.6 aa/turn ∴ 1.5 Å/aa C-term α-Helices have a dipole moment, due to unbonded and aligned N-H and C=O groups β-Sheets contain extended (β-strand) segments from separate regions of a protein prototypical φ = -139 °, ψ = +135 ° prototypical φ = -119 °, ψ = +113 ° (6.5Å repeat length in parallel sheet) Antiparallel β-sheets may be formed by closer regions of sequence than parallel Beta turn Figure 6-13 The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation • Interactions between adjacent R-groups – Ionic attraction or repulsion – Steric hindrance of adjacent bulky groups Helix wheel The stability of helices and sheets depends on their sequence of amino acids • Intrinsic propensity of an amino acid to adopt a helical or extended (strand) conformation • Interactions between adjacent R-groups – Ionic attraction or repulsion – Steric hindrance of adjacent bulky groups • Occurrence of proline and glycine • Interactions between ends of helix and aa R-groups His Glu N-term C-term -

(12) Patent Application Publication (10) Pub. No.: US 2008/0085284 A1 Patell Et Al
US 20080O85284A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2008/0085284 A1 Patell et al. (43) Pub. Date: Apr. 10, 2008 (54) CONSTRUCTION OF A COMPARATIVE (52) U.S. Cl. ............................. 424/184.1; 435/6: 514/44; DATABASE AND IDENTIFICATION OF 536/23.7:536/24.33: 707/102 VIRULENCE FACTORS COMPARISON OF POLYMORPHC REGIONS IN CLINICAL (57) ABSTRACT SOLATES OF INFECTIOUS ORGANISMS The present invention is directed to novel nucleotide (76) Inventors: Villoo Morawala Patell, Bangalore sequences to be used for diagnosis, identification of the (IN); K.R. Rajyashri, Bangalore (IN); strain, typing of the strain and giving orientation to its Marc Rodrigue, Marcy (FR); Guy potential degree of virulence, infectivity and/or latency for Vernet, Marcy (FR) all infectious diseases more particularly tuberculosis. The present invention also includes method for the identification Correspondence Address: and selection of polymorphisms associated with the viru SALWANCHIK LLOYD & SALWANCHK lence and/or infectivity in infectious diseases more particu A PROFESSIONAL ASSOCATION larly in tuberculosis by a comparative genomic analysis of PO BOX 142950 the sequences of different clinical isolates/strains of infec GAINESVILLE, FL 32614-2950 (US) tious organisms. The regions of polymorphisms, can also act (21) Appl. No.: 11/632,108 as potential drug targets and vaccine targets. More particu larly, the invention also relates to identifying virulence (22) Filed: Apr. 9, 2007 factors of M. tuberculosis strains and other infectious organ isms to be included in a diagnostic DNA chip allowing Publication Classification identification of the strain, typing of the strain and finally (51) Int. Cl. -

A Universal Method for Automated Gene Mapping Comment Peder Zipperlen¤*, Knud Nairz¤†, Ivo Rimann†, Konrad Basler*, Ernst Hafen†, Michael Hengartner* and Alex Hajnal†
Open Access Method2005ZipperlenetVolume al. 6, Issue 2, Article R19 A universal method for automated gene mapping comment Peder Zipperlen¤*, Knud Nairz¤†, Ivo Rimann†, Konrad Basler*, Ernst Hafen†, Michael Hengartner* and Alex Hajnal† Addresses: *Institute of Molecular Biology, University of Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland. †Institute of Zoology, University of Zürich, Winterthurerstrasse 190, CH-8057 Zürich, Switzerland. ¤ These authors contributed equally to this work. reviews Correspondence: Peder Zipperlen. E-mail: [email protected]. Knud Nairz. E-mail: [email protected] Published: 17 January 2005 Received: 9 September 2004 Revised: 15 November 2004 Genome Biology 2005, 6:R19 Accepted: 9 December 2004 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2005/6/2/R19 reports © 2005 Zipperlen et al.; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Mapping<p>Amaps forhigh-throughput DrosophilaInDel sequence and method C.polymorphisms. elegans.</p> for genotyping by mapping InDels. This method has been used to create fragment-length polymorphism deposited research Abstract Small insertions or deletions (InDels) constitute a ubiquituous class of sequence polymorphisms found in eukaryotic genomes. Here, we present an automated high-throughput genotyping method that relies on the detection of fragment-length polymorphisms (FLPs) caused by InDels. The protocol utilizes standard sequencers and genotyping software. We have established genome-wide FLP maps for both Caenorhabditis elegans and Drosophila melanogaster that facilitate genetic mapping with a minimum of manual input and at comparatively low cost. -

Adriana Antônia Da Cruz Furini Malária Vivax No Estado
Faculdade de Medicina de São José do Rio Preto Programa de Pós-graduação em Ciências da Saúde Adriana Antônia da Cruz Furini Malária vivax no Estado do Pará: influência de polimorfismos nos genes TNFA, IFNG e IL10 associados à resposta imune humoral e ancestralidade genômica. São José do Rio Preto 2016 Adriana Antônia da Cruz Furini Malária vivax no Estado do Pará: influência de polimorfismos nos genes TNFA, IFNG e IL10 associados à resposta imune humoral e ancestralidade genômica. Tese apresentada à Faculdade de Medicina de São José do Rio Preto para obtenção do Título de Doutor no Programa de Pós Graduação em Ciências da Saúde, Eixo Temático: Medicina e Ciências Correlatas. Orientador: Prof. Dr. Ricardo Luiz Dantas Machado São José do Rio Preto 2016 Furini, Adriana Antônia da Cruz Malária vivax no Estado do Pará: influência de polimorfismos nos genes TNFA, IFNG e IL10 associados à resposta imune humoral e ancestralidade genômica./ Adriana Antônia da Cruz Furini São José do Rio Preto, 2016. 124p. Tese (Doutorado) – Faculdade de Medicina de São José do Rio Preto – FAMERP Eixo Temático: Medicina e Ciências Correlatas Orientador: Prof. Dr. Ricardo Luiz Dantas Machado 1.Ancestralidade; 2. Anticorpos; 3. Citocinas; 4. Malária; 5.Plasmodium vivax. ADRIANA ANTÔNIA DA CRUZ FURINI Malária vivax no Estado do Pará: influência de polimorfismos nos genes TNFA, IFNG e IL10 associados à resposta imune humoral e ancestralidade genômica. BANCA EXAMINADORA TESE PARA OBTENÇÃO DO GRAU DE DOUTOR Presidente/Orientador: Prof. Dr. Ricardo Luiz Dantas Machado 2º Examinador:Prof. Dr. Carlos Eugênio Cavasini 3º Examinador: Profa. Dra. Heloísa da Silveira Paro Pedro 4º Examinador: Profa. -

Synthesis and Nmr Studies of a Β-Turn Mimetic Molecular
SYNTHESIS AND NMR STUDIES OF A β-TURN MIMETIC MOLECULAR TORSION BALANCE by Melissa Ann Liberatore B.S. Chemistry, Lehigh University, 2006 Submitted to the Graduate Faculty of the Kenneth P. Dietrich School of Arts and Sciences in partial fulfillment of the requirements for the degree of Doctor of Philosophy University of Pittsburgh 2012 UNIVERSITY OF PITTSBURGH DIETRICH SCHOOL OF ARTS AND SCIENCES This dissertation was presented by Melissa Ann Liberatore It was defended on July 23, 2012 and approved by Professor Dennis Curran, Department of Chemistry Professor Michael Trakselis, Department of Chemistry Professor Judith Klein-Seetharaman, Department of Structural Biology Dissertation Advisor: Professor Craig Wilcox, Department of Chemistry ii Copyright © by Melissa Ann Liberatore 2012 iii SYNTHESIS AND NMR STUDIES OF A β-TURN MIMETIC MOLECULAR TORSION BALANCE Melissa Ann Liberatore, PhD University of Pittsburgh, 2012 The attainment of precise measurements of the molecular forces that influence protein folding is important in order to further understand peptide dynamics and stability. A hybrid synthetic- natural peptide motif, combining an o,o,o’-trisubstituted biphenyl with an (ortho-tolyl)-amide, was synthesized in multiple formats and studied by NMR to probe the effects of amino acid substitutions on antiparallel beta-sheet configuration and stability. The potential of this “molecular torsion balance” as a beta-turn mimic was demonstrated by quantifying the rotational barriers about several axes. The free-energy rotational barrier of the aryl-aryl bond was found to be 35.7 kcal mol-1 at 418 K in hexanes. EXSY analysis was also used to measure barriers about -1 -1 the N-aryl (20.9 kcal mol at 343 K in toluene-d8) and N-CO bonds (17.2 kcal mol at 298 K in chloroform-d). -

Duke University Dissertation Template
Order and Disorder in Protein Biomaterial Design by Stefan Daniel Roberts Department of Biomedical Engineering Duke University Date:_______________________ Approved: ___________________________ Ashutosh Chilkoti, Supervisor ___________________________ Joel Collier ___________________________ Brenton Hoffman ___________________________ Terrence Oas ___________________________ Rohit Pappu Dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Biomedical Engineering in the Graduate School of Duke University 2018 ABSTRACT Order and Disorder in Protein Biomaterial Design by Stefan Daniel Roberts Department of Biomedical Engineering Duke University Date:_______________________ Approved: ___________________________ Ashutosh Chilkoti, Supervisor ___________________________ Joel Collier ___________________________ Brenton Hoffman ___________________________ Terrence Oas ___________________________ Rohit Pappu An abstract of a dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Department of Biomedical Engineering in the Graduate School of Duke University 2018 Copyright by Stefan Daniel Roberts 2018 Abstract Crystalline and amorphous materials have been extensively studied for their interesting properties, but they comprise a very small portion of the total materials space. The properties of most materials are a consequence of the interactions between their ordered and disordered components. This phenomenon is especially -

(12) United States Patent (10) Patent No.: US 6,686,163 B2 Allen Et Al
US006686163B2 (12) United States Patent (10) Patent No.: US 6,686,163 B2 Allen et al. (45) Date of Patent: Feb. 3, 2004 (54) CODING SEQUENCE HAPLOTYPE OF THE 5,912,127 A 6/1999 Narod et al. ................... 435/6 HUMAN BRCA1 GENE 5.948,643 A 9/1999 Rubinfeld et al. ......... 435/69.1 5,965,377 A 10/1999 Adams et al. ............. 435/7.23 (75) Inventors: Antonette C. P. Allen, Severn, MD 6,033,857 A 3/2000 Tavtigian et al. .............. 435/6 (US); Tracy S. Angelly, Gaithersburg, 6,045,997 A 4/2000 Futreal et al. - - - - - - - - - - 435/6 MD (US); Tammy Lawrence, Laurel, 6,051,379 A 4/2000 Lescallett et al. .............. 435/6 6,083,698 A 7/2000 Olson et al. ................... 435/6 MD (US); Sheri J.Olson, Falls 6,124,104 A 9/2000 Tavtigian et al. ............ 435/7.2 Church, VA (US); Mark B. Rabin, 6,130,322 A * 10/2000 Murphy et al. ................ 435/6 Rockville, MD (US) FOREIGN PATENT DOCUMENTS (73) Assignee: Gene Logic Inc., Gaithersburg, MD EP O705902 A1 4/1996 (US) EP O705903 A1 4/1996 EP O699.754 A1 6/1996 (*) Notice: Subject to any disclaimer, the term of this GB 2307477 A 5/1997 patent is extended or adjusted under 35 WO WO9304200 3/1993 U.S.C. 154(b) by 0 days. WO WO9519369 7/1995 WO WO9722689 6/1997 WO WO973O108 8/1997 (21) Appl. No.: 10/022,819 WO WO9815654 4/1998 (22) Filed: Dec. 20, 2001 OTHER PUBLICATIONS (65) Prior Publication Data Abeliovich et al. -

Transition Bias Influences the Evolution of Antibiotic Resistance in Mycobacterium
bioRxiv preprint doi: https://doi.org/10.1101/421651; this version posted September 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Transition bias influences the evolution of antibiotic resistance in Mycobacterium 2 tuberculosis 3 Joshua L. Payne1,2π*, Fabrizio Menardo3,4 π, Andrej Trauner3,4, Sonia Borrell3,4, Sebastian M. 4 Gygli3,4, Chloe Loiseau3,4, Sebastien Gagneux3,4†, Alex R. Hall1† 5 1. Institute of Integrative Biology, ETH Zurich, Switzerland 6 2. Swiss Institute of Bioinformatics, Lausanne, Switzerland 7 3. Swiss Tropical and Public Health Institute, Basel, Switzerland 8 4. University of Basel, Basel, Switzerland 9 *Correspondence: [email protected] 10 π, These authors contributed equally. 11 †These authors also contributed equally. 12 13 1 bioRxiv preprint doi: https://doi.org/10.1101/421651; this version posted September 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 14 Abstract 15 Transition bias, an overabundance of transitions relative to transversions, has been widely 16 reported among studies of mutations spreading under relaxed selection. However, demonstrating 17 the role of transition bias in adaptive evolution remains challenging. We addressed this challenge 18 by analyzing adaptive antibiotic-resistance mutations in the major human pathogen 19 Mycobacterium tuberculosis.