Isolation of Rickettsia, Rickettsiella, and Spiroplasma from Questing Ticks in Japan Using Arthropod Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hplc-Uv Quantitation of Folate Synthesized by Rickettsia
HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) By Junyan Chen A Thesis Presented to The Faculty of Humboldt State University In Partial Fulfillment of the Requirements for the Degree Master of Science in Biology Committee Membership Dr. Jianmin Zhong, Committee Chair Dr. David S. Baston, Committee Member Dr. Jenny Cappuccio, Committee Member Dr. Jacob Varkey, Committee Member Dr. Erik Jules, Program Graduate Coordinator December 2017 ABSTRACT HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) Junyan Chen Ticks are the most important vector of many infectious diseases in the United States. Understanding the nature of the relationship between Rickettsia endosymbiont Ixodes pacificus (REIP) and Exudes pacificus will help develop strategies for the control of tick- borne diseases, such as Lyme disease, and Rocky Mountain spotted fever. Folate, also known as vitamin B9, is a necessary vitamin for tick survival, and plays a central role in one-carbon metabolism in cells. Folate exist as a large family of structurally related forms that transfer one-carbon groups among biomolecules that are important to cell growth, differentiation, and survival. In Dr. Zheng’s lab, REIP were cultured in Ixodes scapularis embryonic tick cell line ISE6. Previous research has shown that REIP in Ixodes pacificus carries all five de novo folate biosynthesis genes. Folate biosynthesis mRNAs were detected and all recombinant rickettsial folate proteins were overexpressed. To determine whether REIP synthesize folate, we sought to measure the folate concentration in REIP using HPLC-UV quantification with a Diamond HydrideTM liquid chromatography column. 5-methyltetrahydrofolate (5-MTHF), the active circulating form of folate in bacteria was detected. -
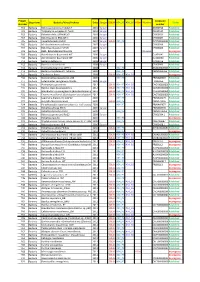
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Diversity of Spotted Fever Group Rickettsiae and Their Association
www.nature.com/scientificreports OPEN Diversity of spotted fever group rickettsiae and their association with host ticks in Japan Received: 31 July 2018 May June Thu1,2, Yongjin Qiu3, Keita Matsuno 4,5, Masahiro Kajihara6, Akina Mori-Kajihara6, Accepted: 14 December 2018 Ryosuke Omori7,8, Naota Monma9, Kazuki Chiba10, Junji Seto11, Mutsuyo Gokuden12, Published: xx xx xxxx Masako Andoh13, Hideo Oosako14, Ken Katakura2, Ayato Takada5,6, Chihiro Sugimoto5,15, Norikazu Isoda1,5 & Ryo Nakao2 Spotted fever group (SFG) rickettsiae are obligate intracellular Gram-negative bacteria mainly associated with ticks. In Japan, several hundred cases of Japanese spotted fever, caused by Rickettsia japonica, are reported annually. Other Rickettsia species are also known to exist in ixodid ticks; however, their phylogenetic position and pathogenic potential are poorly understood. We conducted a nationwide cross-sectional survey on questing ticks to understand the overall diversity of SFG rickettsiae in Japan. Out of 2,189 individuals (19 tick species in 4 genera), 373 (17.0%) samples were positive for Rickettsia spp. as ascertained by real-time PCR amplifcation of the citrate synthase gene (gltA). Conventional PCR and sequencing analyses of gltA indicated the presence of 15 diferent genotypes of SFG rickettsiae. Based on the analysis of fve additional genes, we characterised fve Rickettsia species; R. asiatica, R. helvetica, R. monacensis (formerly reported as Rickettsia sp. In56 in Japan), R. tamurae, and Candidatus R. tarasevichiae and several unclassifed SFG rickettsiae. We also found a strong association between rickettsial genotypes and their host tick species, while there was little association between rickettsial genotypes and their geographical origins. -
Widespread Use of Realtime PCR for Rickettsial Diagnosis
SHORT COMMUNICATION Widespread use of real-time PCR for rickettsial diagnosis Aure´ lie Renvoise´ , Jean-Marc Rolain, Cristina Socolovschi & Didier Raoult Unite´ de Recherche en Maladies Infectieuses et Tropicales Emergentes CNRS-IRD UMR6236-198, Faculte´ de Me´ decine, Universite´ de la Me´ diterrane´ e, Marseille, France Correspondence: Didier Raoult, Unite´ de Abstract Recherche en Maladies Infectieuses et Tropicales Emergentes CNRS-IRD UMR6236- We report 2 years of experience with rickettsial molecular diagnosis using real- 198, Universite´ de la Me´ diterrane´ e, Faculte´ time PCR at the French National Reference Center. All Rickettsia genomes avail- de Me´ decine, 27 bd Jean Moulin, 13385 able were compared to discover specific sequences to design new sets of primers Marseille Cedex 5, France. Tel.: and probes. The specificity was verified in silico and against a panel of 30 rick- +33491324375; fax: +33491387772; ettsial species. Sensitivity was determined using 10-fold serial dilutions. Finally, e-mail: [email protected] primers and probes that were both specific and sensitive were routinely used Received 6 July 2011; revised 6 October for the diagnosis of rickettsial infections from clinical specimens. We retained 2011; accepted 31 October 2011. sets of primers and probes to detect spotted fever group Rickettsia, typhus Final version published online 8 December group Rickettsia, Rickettsia conorii, Rickettsia slovaca, Rickettsia africae and Rick- 2011. ettsia australis; 643 clinical samples were screened for the presence of Rickettsia DNA. Overall, 45 positive samples were detected, including 15 Rickettsia africae, DOI: 10.1111/j.1574-695X.2011.00899.x nine R. conorii, five Rickettsia sibirica mongolitimonae, four R. -

Bacterial Diversity in Amblyomma Americanum (Acari: Ixodidae) with Afocusonmembersofthegenusrickettsia
VECTOR-BORNE DISEASES,SURVEILLANCE,PREVENTION Bacterial Diversity in Amblyomma americanum (Acari: Ixodidae) With aFocusonMembersoftheGenusRickettsia 1 2 1 STEPHANIE R. HEISE, M. S. ELSHAHED, AND S. E. LITTLE Department of Veterinary Pathobiology, Center for Veterinary Health Sciences, Oklahoma State University, Stillwater, OK 74078 J. Med. Entomol. 47(2): 258Ð268 (2010); DOI: 10.1603/ME09197 ABSTRACT The lone star tick, Amblyomma americanum (Acari: Ixodidae), is commonly reported from people and animals throughout the eastern U.S. and is associated with transmission of a number of emerging diseases. To better deÞne the microbial communities within lone star ticks, 16S rRNA gene based analysis using bacteria-wide primers, followed by sequencing of individual clones (n ϭ 449) was used to identify the most common bacterial operational taxonomic units (OTUs) present within colony-reared and wild A. americanum.Thecolony-rearedtickscontainedprimarilysequenceafÞl- iated with members of the genus Coxiella (89%; 81/91), common endosymbionts of ticks, and Brevibacterium (11%; 10/91). Similarly, analysis of clones from unfed wild lone star ticks revealed that 96.7% (89/92) of all the OTUs identiÞed were afÞliated with Coxiella-like endosymbionts, as compared with only 5.1Ð11.7% (5/98Ð9/77) of those identiÞed from wild lone star ticks after feeding. In contrast, the proportion of OTUs identiÞed as Rickettsia sp. in wild-caught ticks increased from 2.2% (2/92) before feeding to as high as 46.8% (36/77) after feeding, and all Rickettsia spp. sequences recovered were most similar to those described from the spotted fever group Rickettsia,speciÞcallyR. amblyo- mmii and R. massiliae.AdditionalcharacterizationoftheRickettsialestickcommunitybypolymerase chain reaction, cloning, and sequencing of 17 kDa and gltA genes conÞrmed these initial Þndings and suggested that novel Rickettsia spp. -

Copyright by Casey L. C. Schroeder 2016
Copyright by Casey L. C. Schroeder 2016 Rickettsia prowazekii and its ‘Junk DNA’: The Identification and Characterization of Small RNAs by Casey Lee Cody Schroeder, MS, MLS(ASCP), SM, SLS Dissertation Presented to the Faculty of the Graduate School of The University of Texas Medical Branch in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy The University of Texas Medical Branch October, 2016 Dedication I dedicate this dissertation to my daughter, who has the fortunate pleasure of possessing half of her mother’s genes. Sorry about the other half. Explore. Dream. Discover. For I learned far more in the woods than I did inside a building. Nature is calling. Answer it. Acknowledgments I would like to acknowledge those that were instrumental: -To the Flying Spaghetti Monster: For your noodly appendage has touched and guided my results. Without the logical conjecture based on overwhelming observable evidence, the conclusions in this dissertation would not have been possible. -The Universe: For had you not expanded 13.8 billion years ago and created the Earth 4.5 billion years ago, none of this research would have been physically possible. The continued spewing of chemically-rich star guts atomically binds this research to the universe and biologically to the Earth. For that, I am eternally grateful. -Dissertation Committee: My sympathies for forcing you to sit through, what probably felt like never-ending, biannual dissertation committee meetings with me. If it is any solace, they have ended………. hopefully. Specifically, Dr. Fofanov, you’re fired. Dr. Minnick, enjoy fly fishing with the grizzly bears. -

Ru 2015 150 263 a (51) Мпк A61k 31/155 (2006.01)
РОССИЙСКАЯ ФЕДЕРАЦИЯ (19) (11) (13) RU 2015 150 263 A (51) МПК A61K 31/155 (2006.01) ФЕДЕРАЛЬНАЯ СЛУЖБА ПО ИНТЕЛЛЕКТУАЛЬНОЙ СОБСТВЕННОСТИ (12) ЗАЯВКА НА ИЗОБРЕТЕНИЕ (21)(22) Заявка: 2015150263, 01.05.2014 (71) Заявитель(и): НЕОКУЛИ ПТИ ЛТД (AU) Приоритет(ы): (30) Конвенционный приоритет: (72) Автор(ы): 01.05.2013 AU 2013901517 ПЕЙДЖ Стефен (AU), ГАРГ Санджай (AU) (43) Дата публикации заявки: 06.06.2017 Бюл. № 16 RU (85) Дата начала рассмотрения заявки PCT на национальной фазе: 01.12.2015 (86) Заявка PCT: AU 2014/000480 (01.05.2014) 2015150263 (87) Публикация заявки PCT: WO 2014/176634 (06.11.2014) Адрес для переписки: 190000, Санкт-Петербург, Box-1125, "ПАТЕНТИКА" A (54) СПОСОБЫ ЛЕЧЕНИЯ БАКТЕРИАЛЬНЫХ ИНФЕКЦИЙ (57) Формула изобретения 1. Способ лечения или профилактики бактериальной колонизации или инфекции у субъекта, включающий стадию: введения субъекту терапевтически эффективного количества робенидина или его терапевтически приемлемой соли, причем указанная A бактериальная колонизация или инфекция вызвана бактериальным агентом. 2. Способ по п. 1, отличающийся тем, что субъект выбран из группы, включающей: человека, животных, принадлежащих видам семейства псовых, кошачьих, крупного рогатого скота, овец, коз, свиней, птиц, рыб и лошадей. 3. Способ по п. 1, отличающийся тем, что робенидин вводят субъекту в дозе в диапазоне от 0,1 до 250 мг/кг массы тела. 4. Способ по любому из пп. 1-3, отличающийся тем, что бактериальный агент является 2015150263 грамположительным. 5. Способ по п. 4, отличающийся тем, что бактериальный агент выбран из -

Pdf 1032003 6
Peer-Reviewed Journal Tracking and Analyzing Disease Trends pages 1891–2100 EDITOR-IN-CHIEF D. Peter Drotman Managing Senior Editor EDITORIAL BOARD Polyxeni Potter, Atlanta, Georgia, USA Dennis Alexander, Addlestone Surrey, United Kingdom Senior Associate Editor Barry J. Beaty, Ft. Collins, Colorado, USA Brian W.J. Mahy, Atlanta, Georgia, USA Martin J. Blaser, New York, New York, USA Christopher Braden, Atlanta, GA, USA Associate Editors Carolyn Bridges, Atlanta, GA, USA Paul Arguin, Atlanta, Georgia, USA Arturo Casadevall, New York, New York, USA Charles Ben Beard, Ft. Collins, Colorado, USA Kenneth C. Castro, Atlanta, Georgia, USA David Bell, Atlanta, Georgia, USA Thomas Cleary, Houston, Texas, USA Charles H. Calisher, Ft. Collins, Colorado, USA Anne DeGroot, Providence, Rhode Island, USA Michel Drancourt, Marseille, France Vincent Deubel, Shanghai, China Paul V. Effl er, Perth, Australia Ed Eitzen, Washington, DC, USA K. Mills McNeill, Kampala, Uganda David Freedman, Birmingham, AL, USA Nina Marano, Atlanta, Georgia, USA Kathleen Gensheimer, Cambridge, MA, USA Martin I. Meltzer, Atlanta, Georgia, USA Peter Gerner-Smidt, Atlanta, GA, USA David Morens, Bethesda, Maryland, USA Duane J. Gubler, Singapore J. Glenn Morris, Gainesville, Florida, USA Richard L. Guerrant, Charlottesville, Virginia, USA Patrice Nordmann, Paris, France Scott Halstead, Arlington, Virginia, USA Tanja Popovic, Atlanta, Georgia, USA David L. Heymann, Geneva, Switzerland Jocelyn A. Rankin, Atlanta, Georgia, USA Daniel B. Jernigan, Atlanta, Georgia, USA Didier Raoult, Marseille, France Charles King, Cleveland, Ohio, USA Pierre Rollin, Atlanta, Georgia, USA Keith Klugman, Atlanta, Georgia, USA Dixie E. Snider, Atlanta, Georgia, USA Takeshi Kurata, Tokyo, Japan Frank Sorvillo, Los Angeles, California, USA S.K. Lam, Kuala Lumpur, Malaysia David Walker, Galveston, Texas, USA Bruce R. -

Clinical Infectious Diseases Emerging Concepts and Strategies in Clinical
Clinical Infectious Diseases Emerging Concepts and Strategies in Clinical Microbiology and Infectious Diseases Guest Editors: Didier Raoult, MD, PhD Fernando Baquero, MD, PhD This supplement was sponsored by the Fondation Méditerranée Infection (FMI). Cover Image: View of the new Méditerranée Infection building in Marseille, France. Photograph by Oleg Mediannikov © Oleg Mediannikov. The opinions expressed in this publication are those of the authors and are not attributable to the sponsors or to the publisher, editor, or editorial board of Clinical Infectious Diseases. Articles may refer to uses of drugs or dosages for periods of time, for indication, or in combinations not included in the current prescribing information. The reader is therefore urged to check the full prescribing information for each drug for the recommended indications, dosage, and precautions and to use clinical judgment in weighing benefits against risk of toxicity. MARCH20176513 15 August 2017 Volume 65 Clinical Infectious Diseases Supplement 1 The title Clinical Infectious Diseases is a registered trademark of the IDSA EMERGING CONCEPTS AND STRATEGIES IN CLINICAL MICROBIOLOGY AND INFECTIOUS DISEASES S1 Rewiring Microbiology and Infection S55 A Hospital-Based Committee of Moral Philosophy Didier Raoult and Fernando Baquero to Revive Ethics Margaux Illy, Pierre Le Coz, and Jean-Louis Mege S4 Building an Intelligent Hospital to Fight Contagion S58 Evaluating the Clinical Burden and Mortality Jérôme Bataille and Philippe Brouqui Attributable to Antibiotic Resistance: -

Appendix 1. Validly Published Names, Conserved and Rejected Names, And
Appendix 1. Validly published names, conserved and rejected names, and taxonomic opinions cited in the International Journal of Systematic and Evolutionary Microbiology since publication of Volume 2 of the Second Edition of the Systematics* JEAN P. EUZÉBY New phyla Alteromonadales Bowman and McMeekin 2005, 2235VP – Valid publication: Validation List no. 106 – Effective publication: Names above the rank of class are not covered by the Rules of Bowman and McMeekin (2005) the Bacteriological Code (1990 Revision), and the names of phyla are not to be regarded as having been validly published. These Anaerolineales Yamada et al. 2006, 1338VP names are listed for completeness. Bdellovibrionales Garrity et al. 2006, 1VP – Valid publication: Lentisphaerae Cho et al. 2004 – Valid publication: Validation List Validation List no. 107 – Effective publication: Garrity et al. no. 98 – Effective publication: J.C. Cho et al. (2004) (2005xxxvi) Proteobacteria Garrity et al. 2005 – Valid publication: Validation Burkholderiales Garrity et al. 2006, 1VP – Valid publication: Vali- List no. 106 – Effective publication: Garrity et al. (2005i) dation List no. 107 – Effective publication: Garrity et al. (2005xxiii) New classes Caldilineales Yamada et al. 2006, 1339VP VP Alphaproteobacteria Garrity et al. 2006, 1 – Valid publication: Campylobacterales Garrity et al. 2006, 1VP – Valid publication: Validation List no. 107 – Effective publication: Garrity et al. Validation List no. 107 – Effective publication: Garrity et al. (2005xv) (2005xxxixi) VP Anaerolineae Yamada et al. 2006, 1336 Cardiobacteriales Garrity et al. 2005, 2235VP – Valid publica- Betaproteobacteria Garrity et al. 2006, 1VP – Valid publication: tion: Validation List no. 106 – Effective publication: Garrity Validation List no. 107 – Effective publication: Garrity et al. -

Update on Tick-Borne Rickettsioses Around the World: a Geographic Approach
Update on Tick-Borne Rickettsioses around the World: a Geographic Approach Philippe Parola,a Christopher D. Paddock,b Cristina Socolovschi,a Marcelo B. Labruna,c Oleg Mediannikov,a Tahar Kernif,d Mohammad Yazid Abdad,e* John Stenos,e Idir Bitam,f Pierre-Edouard Fournier,a Didier Raoulta Aix Marseille Université, Unité de Recherche en Maladies Infectieuses et Tropicales Emergentes (URMITE), UM63, CNRS 7278, IRD 198, Inserm 1095, WHO Collaborative Center for Rickettsioses and Other Arthropod-Borne Bacterial Diseases, Faculté de Médecine, Marseille, Francea; Centers for Disease Control and Prevention, Atlanta, Georgia, USAb; Departamento de Medicina Veterinária Preventiva e Saúde Animal, Faculdade de Medicina Veterinária e Zootecnia Universidade de São Paulo, Cidade Universitária, São Paulo, SP, Brazilc; Service d’Ecologie des Systèmes Vectoriels, Institut Pasteur d’Algérie, Algiers, Algeriad; Division of Veterinary and Biomedical Science, Murdoch University, Australian Rickettsial Reference Laboratory, Barwon Health, Geelong, Victoria, Australiae; University of Boumerdes, Boumerdes, Algeriaf SUMMARY ..................................................................................................................................................658 INTRODUCTION ............................................................................................................................................659 TAXONOMIC AND GENOMIC BACTERIOLOGY: THE REVOLUTION ........................................................................................659 -

The Microbiome of North Sea Copepods
Helgol Mar Res (2013) 67:757–773 DOI 10.1007/s10152-013-0361-4 ORIGINAL ARTICLE The microbiome of North Sea copepods G. Gerdts • P. Brandt • K. Kreisel • M. Boersma • K. L. Schoo • A. Wichels Received: 5 March 2013 / Accepted: 29 May 2013 / Published online: 29 June 2013 Ó Springer-Verlag Berlin Heidelberg and AWI 2013 Abstract Copepods can be associated with different kinds Keywords Bacterial community Á Copepod Á and different numbers of bacteria. This was already shown in Helgoland roads Á North Sea the past with culture-dependent microbial methods or microscopy and more recently by using molecular tools. In our present study, we investigated the bacterial community Introduction of four frequently occurring copepod species, Acartia sp., Temora longicornis, Centropages sp. and Calanus helgo- Marine copepods may constitute up to 80 % of the meso- landicus from Helgoland Roads (North Sea) over a period of zooplankton biomass (Verity and Smetacek 1996). They are 2 years using DGGE (denaturing gradient gel electrophore- key components of the food web as grazers of primary pro- sis) and subsequent sequencing of 16S-rDNA fragments. To duction and as food for higher trophic levels, such as fish complement the PCR-DGGE analyses, clone libraries of (Cushing 1989; Møller and Nielsen 2001). Copepods con- copepod samples from June 2007 to 208 were generated. tribute to the microbial loop (Azam et al. 1983) due to Based on the DGGE banding patterns of the two years sur- ‘‘sloppy feeding’’ (Møller and Nielsen 2001) and the release vey, we found no significant differences between the com- of nutrients and DOM from faecal pellets (Hasegawa et al.