Rickettsia Japonica Strains and Other Closely Related Strains, Including R
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hplc-Uv Quantitation of Folate Synthesized by Rickettsia
HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) By Junyan Chen A Thesis Presented to The Faculty of Humboldt State University In Partial Fulfillment of the Requirements for the Degree Master of Science in Biology Committee Membership Dr. Jianmin Zhong, Committee Chair Dr. David S. Baston, Committee Member Dr. Jenny Cappuccio, Committee Member Dr. Jacob Varkey, Committee Member Dr. Erik Jules, Program Graduate Coordinator December 2017 ABSTRACT HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) Junyan Chen Ticks are the most important vector of many infectious diseases in the United States. Understanding the nature of the relationship between Rickettsia endosymbiont Ixodes pacificus (REIP) and Exudes pacificus will help develop strategies for the control of tick- borne diseases, such as Lyme disease, and Rocky Mountain spotted fever. Folate, also known as vitamin B9, is a necessary vitamin for tick survival, and plays a central role in one-carbon metabolism in cells. Folate exist as a large family of structurally related forms that transfer one-carbon groups among biomolecules that are important to cell growth, differentiation, and survival. In Dr. Zheng’s lab, REIP were cultured in Ixodes scapularis embryonic tick cell line ISE6. Previous research has shown that REIP in Ixodes pacificus carries all five de novo folate biosynthesis genes. Folate biosynthesis mRNAs were detected and all recombinant rickettsial folate proteins were overexpressed. To determine whether REIP synthesize folate, we sought to measure the folate concentration in REIP using HPLC-UV quantification with a Diamond HydrideTM liquid chromatography column. 5-methyltetrahydrofolate (5-MTHF), the active circulating form of folate in bacteria was detected. -

(Batch Learning Self-Organizing Maps), to the Microbiome Analysis of Ticks
Title A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks Nakao, Ryo; Abe, Takashi; Nijhof, Ard M; Yamamoto, Seigo; Jongejan, Frans; Ikemura, Toshimichi; Sugimoto, Author(s) Chihiro The ISME Journal, 7(5), 1003-1015 Citation https://doi.org/10.1038/ismej.2012.171 Issue Date 2013-03 Doc URL http://hdl.handle.net/2115/53167 Type article (author version) File Information ISME_Nakao.pdf Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP A novel approach, based on BLSOMs (Batch Learning Self-Organizing Maps), to the microbiome analysis of ticks Ryo Nakao1,a, Takashi Abe2,3,a, Ard M. Nijhof4, Seigo Yamamoto5, Frans Jongejan6,7, Toshimichi Ikemura2, Chihiro Sugimoto1 1Division of Collaboration and Education, Research Center for Zoonosis Control, Hokkaido University, Kita-20, Nishi-10, Kita-ku, Sapporo, Hokkaido 001-0020, Japan 2Nagahama Institute of Bio-Science and Technology, Nagahama, Shiga 526-0829, Japan 3Graduate School of Science & Technology, Niigata University, 8050, Igarashi 2-no-cho, Nishi- ku, Niigata 950-2181, Japan 4Institute for Parasitology and Tropical Veterinary Medicine, Freie Universität Berlin, Königsweg 67, 14163 Berlin, Germany 5Miyazaki Prefectural Institute for Public Health and Environment, 2-3-2 Gakuen Kibanadai Nishi, Miyazaki 889-2155, Japan 6Utrecht Centre for Tick-borne Diseases (UCTD), Department of Infectious Diseases and Immunology, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 1, 3584 CL Utrecht, The Netherlands 7Department of Veterinary Tropical Diseases, Faculty of Veterinary Science, University of Pretoria, Private Bag X04, 0110 Onderstepoort, South Africa aThese authors contributed equally to this work. Keywords: BLSOMs/emerging diseases/metagenomics/microbiomes/symbionts/ticks Running title: Tick microbiomes revealed by BLSOMs Subject category: Microbe-microbe and microbe-host interactions Abstract Ticks transmit a variety of viral, bacterial and protozoal pathogens, which are often zoonotic. -

Transition of Serum Cytokine Concentration in Rickettsia Japonica Infection
Article Transition of Serum Cytokine Concentration in Rickettsia japonica Infection Makoto Kondo, Yoshiaki Matsushima, Kento Mizutani, Shohei Iida, Koji Habe and Keiichi Yamanaka * Department of Dermatology, Mie University Graduate School of medicine, 2-174 Edobashi, Tsu, Mie 514-8507, Japan; [email protected] (M.K.); [email protected] (Y.M.); [email protected] (K.M.); [email protected] (S.I.); [email protected] (K.H.) * Correspondence: [email protected]; Tel.: +81-59-231-5025; Fax: +81-59-231-5206 Received: 31 October 2020; Accepted: 8 December 2020; Published: 11 December 2020 Abstract: (1) Background. Rickettsia japonica (R. japonica) infection induces severe inflammation, and the disappearance of eosinophil in the acute stage is one of the phenomena. (2) Materials and Methods. In the current study, we measured the serum concentrations of Th1, Th2, and Th17 cytokines in the acute and recovery stages. (3) Results. In the acute phase, IL-6 and IFN-γ levels were elevated and we speculated that they played a role as a defense mechanism against R. japonica. The high concentration of IFN-γ suppressed the differentiation of eosinophil and induced apoptosis of eosinophil, leading to the disappearance of eosinophil. On day 7, IL-6 and IFN-γ concentrations were decreased, and Th2 cytokines such as IL-5 and IL-9 were slightly increased. On day 14, eosinophil count recovered to the normal level. The transition of serum cytokine concentration in R. japonica infection was presented. (4) Conclusions. -

Ohio Department of Health, Bureau of Infectious Diseases Disease Name Class A, Requires Immediate Phone Call to Local Health
Ohio Department of Health, Bureau of Infectious Diseases Reporting specifics for select diseases reportable by ELR Class A, requires immediate phone Susceptibilities specimen type Reportable test name (can change if Disease Name other specifics+ call to local health required* specifics~ state/federal case definition or department reporting requirements change) Culture independent diagnostic tests' (CIDT), like BioFire panel or BD MAX, E. histolytica Stain specimen = stool, bile results should be sent as E. histolytica DNA fluid, duodenal fluid, 260373001^DETECTED^SCT with E. histolytica Antigen Amebiasis (Entamoeba histolytica) No No tissue large intestine, disease/organism-specific DNA LOINC E. histolytica Antibody tissue small intestine codes OR a generic CIDT-LOINC code E. histolytica IgM with organism-specific DNA SNOMED E. histolytica IgG codes E. histolytica Total Antibody Ova and Parasite Anthrax Antibody Anthrax Antigen Anthrax EITB Acute Anthrax EITB Convalescent Anthrax Yes No Culture ELISA PCR Stain/microscopy Stain/spore ID Eastern Equine Encephalitis virus Antibody Eastern Equine Encephalitis virus IgG Antibody Eastern Equine Encephalitis virus IgM Arboviral neuroinvasive and non- Eastern Equine Encephalitis virus RNA neuroinvasive disease: Eastern equine California serogroup virus Antibody encephalitis virus disease; LaCrosse Equivocal results are accepted for all California serogroup virus IgG Antibody virus disease (other California arborviral diseases; California serogroup virus IgM Antibody specimen = blood, serum, serogroup -

A Streamlined Method for Transposon Mutagenesis of Rickettsia Parkeri
bioRxiv preprint doi: https://doi.org/10.1101/277160; this version posted March 8, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 A streamlined method for transposon mutagenesis of 2 Rickettsia parkeri yields numerous mutations that 3 impact infection 4 5 Rebecca L. Lamason1,#a,*, Natasha M. Kafai1,#b, and Matthew D. Welch1* 6 7 8 1 Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, 9 CA 10 #a Current address: Department of Biology, Massachusetts Institute of Technology, 11 Cambridge, MA 12 #b Current address: Medical Scientist Training Program, Washington University in St. 13 Louis School of Medicine, St. Louis, MO 14 15 16 17 18 * Co-corresponding authors 19 E-mail: [email protected], [email protected] 20 1 bioRxiv preprint doi: https://doi.org/10.1101/277160; this version posted March 8, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 21 Abstract 22 The rickettsiae are obligate intracellular alphaproteobacteria that exhibit a complex 23 infectious life cycle in both arthropod and mammalian hosts. As obligate intracellular 24 bacteria, Rickettsia are highly adapted to living inside a variety of host cells, including 25 vascular endothelial cells during mammalian infection. Although it is assumed that the 26 rickettsiae produce numerous virulence factors that usurp or disrupt various host cell 27 pathways, they have been challenging to genetically manipulate to identify the key 28 bacterial factors that contribute to infection. -

Burkholderia Lata Infections from Intrinsically Contaminated
RESEARCH LETTERS China need to become aware of R. japonica disease presen- Burkholderia lata Infections tation, so they can administer the appropriate treatment to patients with suspected R. japonica infections. from Intrinsically Contaminated Chlorhexidine This study was supported by the National Natural Science Foundation of China (81571963); Science Foundation of Anhui Mouthwash, Australia, 2016 Province of China (1608085MH213); Natural Science Foundation Key Project of Anhui Province Education Department Lex E.X. Leong, Diana Lagana, Glen P. Carter, (KJ2015A020, KJ2016A331); and Scientific Research of Anhui Qinning Wang, Kija Smith, Tim P. Stinear, Medical University (XJ201314, XJ201430, XJ201503). David Shaw, Vitali Sintchenko, Steven L. Wesselingh, Ivan Bastian, About the Authors Geraint B. Rogers Dr. Li is a research coordinator at The First Affiliated Hospital of Author affiliations: South Australian Health and Medical Research Anhui Medical University, Hefei, China. His research interests Institute, Adelaide, South Australia, Australia (L.E.X. Leong, are pathogenic mechanisms of tickborne infectious diseases, S.L. Wesselingh, G.B. Rogers); Flinders University, Bedford Park, including severe fever with thrombocytopenia syndrome, human South Australia, Australia (L.E.X. Leong, G.B. Rogers); Royal granulocytic anaplasmosis, and spotted fever group rickettsioses. Adelaide Hospital, Adelaide (D. Lagana, D. Shaw); University of Dr. Wen Hu is an electron microscope technician at The First Melbourne, Melbourne, Victoria, Australia (G.P. Carter, Affiliated Hospital of the University of Science and Technology of T.P. Stinear); The University of Sydney, Westmead, New South China, Hefei, China. His research interest is pathogen structure. Wales, Australia (Q. Wang, V. Sintchenko); SA Pathology, Adelaide (K. Smith, I. Bastian) References DOI: https://doi.org/10.3201/eid2411.171929 1. -
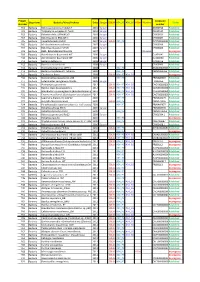
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Diversity of Spotted Fever Group Rickettsiae and Their Association
www.nature.com/scientificreports OPEN Diversity of spotted fever group rickettsiae and their association with host ticks in Japan Received: 31 July 2018 May June Thu1,2, Yongjin Qiu3, Keita Matsuno 4,5, Masahiro Kajihara6, Akina Mori-Kajihara6, Accepted: 14 December 2018 Ryosuke Omori7,8, Naota Monma9, Kazuki Chiba10, Junji Seto11, Mutsuyo Gokuden12, Published: xx xx xxxx Masako Andoh13, Hideo Oosako14, Ken Katakura2, Ayato Takada5,6, Chihiro Sugimoto5,15, Norikazu Isoda1,5 & Ryo Nakao2 Spotted fever group (SFG) rickettsiae are obligate intracellular Gram-negative bacteria mainly associated with ticks. In Japan, several hundred cases of Japanese spotted fever, caused by Rickettsia japonica, are reported annually. Other Rickettsia species are also known to exist in ixodid ticks; however, their phylogenetic position and pathogenic potential are poorly understood. We conducted a nationwide cross-sectional survey on questing ticks to understand the overall diversity of SFG rickettsiae in Japan. Out of 2,189 individuals (19 tick species in 4 genera), 373 (17.0%) samples were positive for Rickettsia spp. as ascertained by real-time PCR amplifcation of the citrate synthase gene (gltA). Conventional PCR and sequencing analyses of gltA indicated the presence of 15 diferent genotypes of SFG rickettsiae. Based on the analysis of fve additional genes, we characterised fve Rickettsia species; R. asiatica, R. helvetica, R. monacensis (formerly reported as Rickettsia sp. In56 in Japan), R. tamurae, and Candidatus R. tarasevichiae and several unclassifed SFG rickettsiae. We also found a strong association between rickettsial genotypes and their host tick species, while there was little association between rickettsial genotypes and their geographical origins. -
Widespread Use of Realtime PCR for Rickettsial Diagnosis
SHORT COMMUNICATION Widespread use of real-time PCR for rickettsial diagnosis Aure´ lie Renvoise´ , Jean-Marc Rolain, Cristina Socolovschi & Didier Raoult Unite´ de Recherche en Maladies Infectieuses et Tropicales Emergentes CNRS-IRD UMR6236-198, Faculte´ de Me´ decine, Universite´ de la Me´ diterrane´ e, Marseille, France Correspondence: Didier Raoult, Unite´ de Abstract Recherche en Maladies Infectieuses et Tropicales Emergentes CNRS-IRD UMR6236- We report 2 years of experience with rickettsial molecular diagnosis using real- 198, Universite´ de la Me´ diterrane´ e, Faculte´ time PCR at the French National Reference Center. All Rickettsia genomes avail- de Me´ decine, 27 bd Jean Moulin, 13385 able were compared to discover specific sequences to design new sets of primers Marseille Cedex 5, France. Tel.: and probes. The specificity was verified in silico and against a panel of 30 rick- +33491324375; fax: +33491387772; ettsial species. Sensitivity was determined using 10-fold serial dilutions. Finally, e-mail: [email protected] primers and probes that were both specific and sensitive were routinely used Received 6 July 2011; revised 6 October for the diagnosis of rickettsial infections from clinical specimens. We retained 2011; accepted 31 October 2011. sets of primers and probes to detect spotted fever group Rickettsia, typhus Final version published online 8 December group Rickettsia, Rickettsia conorii, Rickettsia slovaca, Rickettsia africae and Rick- 2011. ettsia australis; 643 clinical samples were screened for the presence of Rickettsia DNA. Overall, 45 positive samples were detected, including 15 Rickettsia africae, DOI: 10.1111/j.1574-695X.2011.00899.x nine R. conorii, five Rickettsia sibirica mongolitimonae, four R. -

Tick- and Flea-Borne Rickettsial Emerging Zoonoses Philippe Parola, Bernard Davoust, Didier Raoult
Tick- and flea-borne rickettsial emerging zoonoses Philippe Parola, Bernard Davoust, Didier Raoult To cite this version: Philippe Parola, Bernard Davoust, Didier Raoult. Tick- and flea-borne rickettsial emerging zoonoses. Veterinary Research, BioMed Central, 2005, 36 (3), pp.469-492. 10.1051/vetres:2005004. hal- 00902973 HAL Id: hal-00902973 https://hal.archives-ouvertes.fr/hal-00902973 Submitted on 1 Jan 2005 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Vet. Res. 36 (2005) 469–492 469 © INRA, EDP Sciences, 2005 DOI: 10.1051/vetres:2005004 Review article Tick- and flea-borne rickettsial emerging zoonoses Philippe PAROLAa, Bernard DAVOUSTb, Didier RAOULTa* a Unité des Rickettsies, CNRS UMR 6020, IFR 48, Faculté de Médecine, Université de la Méditerranée, 13385 Marseille Cedex 5, France b Direction Régionale du Service de Santé des Armées, BP 16, 69998 Lyon Armées, France (Received 30 March 2004; accepted 5 August 2004) Abstract – Between 1984 and 2004, nine more species or subspecies of spotted fever rickettsiae were identified as emerging agents of tick-borne rickettsioses throughout the world. Six of these species had first been isolated from ticks and later found to be pathogenic to humans. -

Using Core Genome Alignments to Assign Bacterial Species 2
bioRxiv preprint doi: https://doi.org/10.1101/328021; this version posted May 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 1 Using Core Genome Alignments to Assign Bacterial Species 2 3 Matthew Chunga,b, James B. Munroa, Julie C. Dunning Hotoppa,b,c,# 4 a Institute for Genome Sciences, University of Maryland Baltimore, Baltimore, MD 21201, USA 5 b Department of Microbiology and Immunology, University of Maryland Baltimore, Baltimore, MD 6 21201, USA 7 c Greenebaum Comprehensive Cancer Center, University of Maryland Baltimore, Baltimore, MD 21201, 8 USA 9 10 Running Title: Core Genome Alignments to Assign Bacterial Species 11 12 #Address correspondence to Julie C. Dunning Hotopp, [email protected]. 13 14 Word count Abstract: 371 words 15 Word count Text: 4,833 words 16 1 bioRxiv preprint doi: https://doi.org/10.1101/328021; this version posted May 22, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. 17 ABSTRACT 18 With the exponential increase in the number of bacterial taxa with genome sequence data, a new 19 standardized method is needed to assign bacterial species designations using genomic data that is 20 consistent with the classically-obtained taxonomy. -

Bacterial Diversity in Amblyomma Americanum (Acari: Ixodidae) with Afocusonmembersofthegenusrickettsia
VECTOR-BORNE DISEASES,SURVEILLANCE,PREVENTION Bacterial Diversity in Amblyomma americanum (Acari: Ixodidae) With aFocusonMembersoftheGenusRickettsia 1 2 1 STEPHANIE R. HEISE, M. S. ELSHAHED, AND S. E. LITTLE Department of Veterinary Pathobiology, Center for Veterinary Health Sciences, Oklahoma State University, Stillwater, OK 74078 J. Med. Entomol. 47(2): 258Ð268 (2010); DOI: 10.1603/ME09197 ABSTRACT The lone star tick, Amblyomma americanum (Acari: Ixodidae), is commonly reported from people and animals throughout the eastern U.S. and is associated with transmission of a number of emerging diseases. To better deÞne the microbial communities within lone star ticks, 16S rRNA gene based analysis using bacteria-wide primers, followed by sequencing of individual clones (n ϭ 449) was used to identify the most common bacterial operational taxonomic units (OTUs) present within colony-reared and wild A. americanum.Thecolony-rearedtickscontainedprimarilysequenceafÞl- iated with members of the genus Coxiella (89%; 81/91), common endosymbionts of ticks, and Brevibacterium (11%; 10/91). Similarly, analysis of clones from unfed wild lone star ticks revealed that 96.7% (89/92) of all the OTUs identiÞed were afÞliated with Coxiella-like endosymbionts, as compared with only 5.1Ð11.7% (5/98Ð9/77) of those identiÞed from wild lone star ticks after feeding. In contrast, the proportion of OTUs identiÞed as Rickettsia sp. in wild-caught ticks increased from 2.2% (2/92) before feeding to as high as 46.8% (36/77) after feeding, and all Rickettsia spp. sequences recovered were most similar to those described from the spotted fever group Rickettsia,speciÞcallyR. amblyo- mmii and R. massiliae.AdditionalcharacterizationoftheRickettsialestickcommunitybypolymerase chain reaction, cloning, and sequencing of 17 kDa and gltA genes conÞrmed these initial Þndings and suggested that novel Rickettsia spp.