Clinical Infectious Diseases Emerging Concepts and Strategies in Clinical
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hplc-Uv Quantitation of Folate Synthesized by Rickettsia
HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) By Junyan Chen A Thesis Presented to The Faculty of Humboldt State University In Partial Fulfillment of the Requirements for the Degree Master of Science in Biology Committee Membership Dr. Jianmin Zhong, Committee Chair Dr. David S. Baston, Committee Member Dr. Jenny Cappuccio, Committee Member Dr. Jacob Varkey, Committee Member Dr. Erik Jules, Program Graduate Coordinator December 2017 ABSTRACT HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) Junyan Chen Ticks are the most important vector of many infectious diseases in the United States. Understanding the nature of the relationship between Rickettsia endosymbiont Ixodes pacificus (REIP) and Exudes pacificus will help develop strategies for the control of tick- borne diseases, such as Lyme disease, and Rocky Mountain spotted fever. Folate, also known as vitamin B9, is a necessary vitamin for tick survival, and plays a central role in one-carbon metabolism in cells. Folate exist as a large family of structurally related forms that transfer one-carbon groups among biomolecules that are important to cell growth, differentiation, and survival. In Dr. Zheng’s lab, REIP were cultured in Ixodes scapularis embryonic tick cell line ISE6. Previous research has shown that REIP in Ixodes pacificus carries all five de novo folate biosynthesis genes. Folate biosynthesis mRNAs were detected and all recombinant rickettsial folate proteins were overexpressed. To determine whether REIP synthesize folate, we sought to measure the folate concentration in REIP using HPLC-UV quantification with a Diamond HydrideTM liquid chromatography column. 5-methyltetrahydrofolate (5-MTHF), the active circulating form of folate in bacteria was detected. -

Community Analysis of Microbial Sharing and Specialization in A
Downloaded from http://rspb.royalsocietypublishing.org/ on March 15, 2017 Community analysis of microbial sharing rspb.royalsocietypublishing.org and specialization in a Costa Rican ant–plant–hemipteran symbiosis Elizabeth G. Pringle1,2 and Corrie S. Moreau3 Research 1Department of Biology, Program in Ecology, Evolution, and Conservation Biology, University of Nevada, Cite this article: Pringle EG, Moreau CS. 2017 Reno, NV 89557, USA 2Michigan Society of Fellows, University of Michigan, Ann Arbor, MI 48109, USA Community analysis of microbial sharing and 3Department of Science and Education, Field Museum of Natural History, 1400 South Lake Shore Drive, specialization in a Costa Rican ant–plant– Chicago, IL 60605, USA hemipteran symbiosis. Proc. R. Soc. B 284: EGP, 0000-0002-4398-9272 20162770. http://dx.doi.org/10.1098/rspb.2016.2770 Ants have long been renowned for their intimate mutualisms with tropho- bionts and plants and more recently appreciated for their widespread and diverse interactions with microbes. An open question in symbiosis research is the extent to which environmental influence, including the exchange of Received: 14 December 2016 microbes between interacting macroorganisms, affects the composition and Accepted: 17 January 2017 function of symbiotic microbial communities. Here we approached this ques- tion by investigating symbiosis within symbiosis. Ant–plant–hemipteran symbioses are hallmarks of tropical ecosystems that produce persistent close contact among the macroorganism partners, which then have substantial opportunity to exchange symbiotic microbes. We used metabarcoding and Subject Category: quantitative PCR to examine community structure of both bacteria and Ecology fungi in a Neotropical ant–plant–scale-insect symbiosis. Both phloem-feed- ing scale insects and honeydew-feeding ants make use of microbial Subject Areas: symbionts to subsist on phloem-derived diets of suboptimal nutritional qual- ecology, evolution, microbiology ity. -

Case Report: Coinfection with Rickettsia Monacensis and Orientia Tsutsugamushi
Am. J. Trop. Med. Hyg., 101(2), 2019, pp. 332–335 doi:10.4269/ajtmh.18-0631 Copyright © 2019 by The American Society of Tropical Medicine and Hygiene Case Report: Coinfection with Rickettsia monacensis and Orientia tsutsugamushi Seok Won Kim,1† Choon-Mee Kim,2† Dong-Min Kim,3* and Na Ra Yun3 1Department of Neurosurgery, College of Medicine, Chosun University, Gwangju, Republic of Korea; 2Premedical Science, College of Medicine, Chosun University, Gwangju, Republic of Korea; 3Department of Internal Medicine, College of Medicine, Chosun University, Gwangju, Republic of Korea Abstract. Rickettsia monacensis and Orientia tsutsugamushi are bacteria of the family Rickettsiaceae, which causes fever, rash, and eschar formation; outdoor activities are a risk factor for Rickettsiaceae infection. A 75-year-old woman presented with fever, rash, and eschar and was confirmed as being scrub typhus based on a nested-polymerase chain reaction (N-PCR) test for a 56-kDa gene of O. tsutsugamushi; the genome was identified as the Boryong genotype. In addition, a pan-Rickettsia real-time PCR test was positive and a N-PCR test using a Rickettsia-specific partial outer membrane protein A (rOmpA) confirmed R. monacensis. This is the first case wherein a patient suspected of having scrub typhus owing to the presence of rash and eschar was also found to be coinfected with O. tsutsugamushi and R. monacensis based on molecular testing. INTRODUCTION leukocyte count, 7,200/mm3; hemoglobin, 11.6 g/dL; platelet count, 232,000/mm3; and erythrocyte sedimentation rate, 31 Rickettsia monacensis is a pathogen that causes spotted mm/hours. C-reactive protein and procalcitonin levels were fever group rickettsial infection; the main symptoms of in- elevated at 9.26 mg/dL and 0.836 ng/mL (0–0.5 ng/mL), re- fection include fever, headache, and myalgia, as well as es- 1 spectively. -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

(Chiroptera: Vespertilionidae) and the Bat Soft Tick Argas Vespe
Zhao et al. Parasites Vectors (2020) 13:10 https://doi.org/10.1186/s13071-020-3885-x Parasites & Vectors SHORT REPORT Open Access Rickettsiae in the common pipistrelle Pipistrellus pipistrellus (Chiroptera: Vespertilionidae) and the bat soft tick Argas vespertilionis (Ixodida: Argasidae) Shuo Zhao1†, Meihua Yang2†, Gang Liu1†, Sándor Hornok3, Shanshan Zhao1, Chunli Sang1, Wenbo Tan1 and Yuanzhi Wang1* Abstract Background: Increasing molecular evidence supports that bats and/or their ectoparasites may harbor vector-borne bacteria, such as bartonellae and borreliae. However, the simultaneous occurrence of rickettsiae in bats and bat ticks has been poorly studied. Methods: In this study, 54 bat carcasses and their infesting soft ticks (n 67) were collected in Shihezi City, north- western China. The heart, liver, spleen, lung, kidney, small intestine and large= intestine of bats were dissected, followed by DNA extraction. Soft ticks were identifed both morphologically and molecularly. All samples were examined for the presence of rickettsiae by amplifying four genetic markers (17-kDa, gltA, ompA and ompB). Results: All bats were identifed as Pipistrellus pipistrellus, and their ticks as Argas vespertilionis. Molecular analyses showed that DNA of Rickettsia parkeri, R. lusitaniae, R. slovaca and R. raoultii was present in bat organs/tissues. In addition, nine of the 67 bat soft ticks (13.43%) were positive for R. raoultii (n 5) and R. rickettsii (n 4). In the phylo- genetic analysis, these bat-associated rickettsiae clustered together with conspecifc= sequences reported= from other host and tick species, confrming the above results. Conclusions: To the best of our knowledge, DNA of R. parkeri, R. slovaca and R. -

Detection and Partial Molecular Characterization of Rickettsia and Bartonella from Southern African Bat Species
Detection and partial molecular characterization of Rickettsia and Bartonella from southern African bat species by Tjale Mabotse Augustine (29685690) Submitted in partial fulfillment of the requirements for the degree MAGISTER SCIENTIAE (MICROBIOLOGY) in the Department of Microbiology and Plant Pathology Faculty of Natural and Agricultural Sciences University of Pretoria Pretoria, South Africa Supervisor: Dr Wanda Markotter Co-supervisors: Prof Louis H. Nel Dr Jacqueline Weyer May, 2012 I declare that the thesis, which I hereby submit for the degree MSc (Microbiology) at the University of Pretoria, South Africa, is my own work and has not been submitted by me for a degree at another university ________________________________ Tjale Mabotse Augustine i Acknowledgements I would like send my sincere gratitude to the following people: Dr Wanda Markotter (University of Pretoria), Dr Jacqueline Weyer (National Institute for Communicable Diseases-National Health Laboratory Service) and Prof Louis H Nel (University of Pretoria) for their supervision and guidance during the project. Dr Jacqueline Weyer (Centre for Zoonotic and Emerging diseases (Previously Special Pathogens Unit), National Institute for Communicable Diseases (National Heath Laboratory Service), for providing the positive control DNA for Rickettsia and Dr Jenny Rossouw (Special Bacterial Pathogens Reference Unit, National Institute for Communicable Diseases-National Health Laboratory Service), for providing the positive control DNA for Bartonella. Dr Teresa Kearney (Ditsong Museum of Natural Science), Gauteng and Northern Region Bat Interest Group, Kwa-Zulu Natal Bat Interest Group, Prof Ara Monadjem (University of Swaziland), Werner Marias (University of Johannesburg), Dr Francois du Rand (University of Johannesburg) and Prof David Jacobs (University of Cape Town) for collection of blood samples. -

Human Case of Bartonella Alsatica Lymphadenitis
LETTERS (6). The sequence is distinct from a DOI: 10.3201/eid1412.080944 Human Case of small number of sequences derived from rabies viruses in Vietnam, which References Bartonella alsatica suggests that China is a stronger can- Lymphadenitis 1. Smith JS, Fishbein DB, Rupprecht CE, didate for the source of the virus than Clark K. Unexplained rabies in three To the Editor: Lymph node en- her native country. immigrants in the United States: a vi- Although the case history could rologic investigation. N Engl J Med. largement is a common medical prob- not provide evidence for interaction 1991;324:205–11. lem that is usually caused by bacterial, 2. Grattan-Smith PJ, O’Regan WJ, Ellis PS, with a dog while her family was in viral, fungal, or protozoal agents (1). O’Flaherty SJ, McIntyre PB, Barnes CJ. A Malignancies or lymphoproliferative Hong Kong Special Administrative second Australian case, with a long incuba- Region, rabies was endemic within tion period. Med J Aust. 1992;156:651–4. diseases are often found, especially in the colony at the time that the pa- 3. McColl KA, Gould AR, Selleck PW, elderly patients (1). Bartonella hense- Hooper PT, Westbury HA, Smith JS. tient’s family was resident. From 1980 lae, the main causative agent of cat- Polymerase chain reaction and other labo- scratch disease (CSD), appears to be through 1984, 5 human cases were re- ratory techniques in the diagnosis of long corded (9). Only 2 case-patients had incubation rabies in Australia. Aust Vet the most common organism respon- clear evidence of a dog bite; histories J. -

Expansion of Tick-Borne Rickettsioses in the World
microorganisms Review Expansion of Tick-Borne Rickettsioses in the World Mariusz Piotrowski * and Anna Rymaszewska Institute of Biology, University of Szczecin, 70-453 Szczecin, Poland; [email protected] * Correspondence: [email protected] Received: 24 September 2020; Accepted: 25 November 2020; Published: 30 November 2020 Abstract: Tick-borne rickettsioses are caused by obligate intracellular bacteria belonging to the spotted fever group of the genus Rickettsia. These infections are among the oldest known diseases transmitted by vectors. In the last three decades there has been a rapid increase in the recognition of this disease complex. This unusual expansion of information was mainly caused by the development of molecular diagnostic techniques that have facilitated the identification of new and previously recognized rickettsiae. A lot of currently known bacteria of the genus Rickettsia have been considered nonpathogenic for years, and moreover, many new species have been identified with unknown pathogenicity. The genus Rickettsia is distributed all over the world. Many Rickettsia species are present on several continents. The geographical distribution of rickettsiae is related to their vectors. New cases of rickettsioses and new locations, where the presence of these bacteria is recognized, are still being identified. The variety and rapid evolution of the distribution and density of ticks and diseases which they transmit shows us the scale of the problem. This review article presents a comparison of the current understanding of the geographic distribution of pathogenic Rickettsia species to that of the beginning of the century. Keywords: Tick-borne rickettsioses; Tick-borne diseases; Rickettsiales 1. Introduction Tick-borne rickettsioses are caused by obligate intracellular Gram-negative bacteria belonging to the spotted fever group (SFG) of the genus Rickettsia. -

Rickettsia Monacensis As a Cause of a Tick Bite
a nonpruritic, disseminated maculopapular rash, with no Rickettsia inoculation eschar, of the trunk and lower extremities, in- cluding palms and soles. Other than a slightly low plate- monacensis let count (82,000/mm3), examination fi ndings were within normal limits. MSF was diagnosed, and serum and defi - and Human brinated blood samples were taken before a course of oral doxycycline (100 mg/12 h for 10 d) was initiated. Three Disease, Spain days later, fever and rash were gone without sequelae. Ad- Isabel Jado,* José A. Oteo,† Mikel Aldámiz,‡ ditional serial serum samples were taken during weeks 4, Horacio Gil,* Raquel Escudero,* 13, and 26 after onset and reserved for serologic analysis Valvanera Ibarra,† Joseba Portu,‡ (Table). Aranzazu Portillo,† María J. Lezaun,‡ Patient 2 was a 59-year-old woman from Basque Cristina García-Amil,* Isabel Rodríguez-Moreno,* Country, who sought medical attention on September 20, and Pedro Anda* 2003, 4 days after onset of fever (38ºC), headache, and an erythematous rash, with no inoculation eschar, at the site of We identifi ed Rickettsia monacensis as a cause of a tick bite. The patient reported a history of tick bites, most acute tickborne rickettsiosis in 2 humans. Its pathogenic recently 1 week before symptom onset. Blood cell counts role was assessed by culture and detection of the organism and other blood chemistry values were normal. MSF was in patients’ blood samples. This fi nding increases the num- ber of recognized human rickettsial pathogens and expands diagnosed, and oral doxycycline (100 mg/12 h for 10 d) the known geographic distribution of Mediterranean spotted was prescribed. -

Intraspecies Comparative Genomics of Rickettsia
AIX ͲMARSEILLE UNIVERSITÉ FACULTÉ DE MÉDECINE DE MARSEILLE ÉCOLE DOCTORALE DES SCIENCES DE LA VIE ET DE LA SANTÉ T H È S E Présentée et publiquement soutenue devant LA FACULTÉ DE MÉDECINE DE MARSEILLE Le 13 décembre 2013 Par M. Erwin SENTAUSA Né le 16 décembre 1979 àMalang, Indonésie INTRASPECIES COMPARATIVE GENOMICS OF RICKETTSIA Pour obtenir le grade de DOCTORAT d’AIX ͲMARSEILLE UNIVERSITÉ SPÉCIALITÉ :PATHOLOGIE HUMAINE Ͳ MALADIES INFECTIEUSES Membres du Jury de la Thèse : Dr. Patricia RENESTO Rapporteur Pr. Max MAURIN Rapporteur Dr. Florence FENOLLAR Membre du Jury Pr. Pierre ͲEdouard FOURNIER Directeur de thèse Unité de Recherche sur les Maladies Infectieuses et Tropicales Émergentes UM63, CNRS 7278, IRD 198, Inserm 1095 Avant Propos Le format de présentation de cette thèse correspond à une recommandation de la spécialité Maladies Infectieuses et Microbiologie, à l’intérieur du Master de Sciences de la Vie et de la Santé qui dépend de l’Ecole Doctorale des Sciences de la Vie de Marseille. Le candidat est amené àrespecter des règles qui lui sont imposées et qui comportent un format de thèse utilisé dans le Nord de l’Europe permettant un meilleur rangement que les thèses traditionnelles. Par ailleurs, la partie introduction et bibliographie est remplacée par une revue envoyée dans un journal afin de permettre une évaluation extérieure de la qualité de la revue et de permettre àl’étudiant de le commencer le plus tôt possible une bibliographie exhaustive sur le domaine de cette thèse. Par ailleurs, la thèse est présentée sur article publié, accepté ou soumis associé d’un bref commentaire donnant le sens général du travail. -
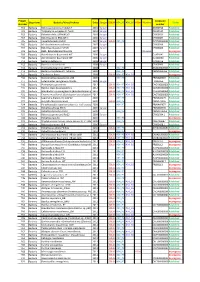
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Diversity of Spotted Fever Group Rickettsiae and Their Association
www.nature.com/scientificreports OPEN Diversity of spotted fever group rickettsiae and their association with host ticks in Japan Received: 31 July 2018 May June Thu1,2, Yongjin Qiu3, Keita Matsuno 4,5, Masahiro Kajihara6, Akina Mori-Kajihara6, Accepted: 14 December 2018 Ryosuke Omori7,8, Naota Monma9, Kazuki Chiba10, Junji Seto11, Mutsuyo Gokuden12, Published: xx xx xxxx Masako Andoh13, Hideo Oosako14, Ken Katakura2, Ayato Takada5,6, Chihiro Sugimoto5,15, Norikazu Isoda1,5 & Ryo Nakao2 Spotted fever group (SFG) rickettsiae are obligate intracellular Gram-negative bacteria mainly associated with ticks. In Japan, several hundred cases of Japanese spotted fever, caused by Rickettsia japonica, are reported annually. Other Rickettsia species are also known to exist in ixodid ticks; however, their phylogenetic position and pathogenic potential are poorly understood. We conducted a nationwide cross-sectional survey on questing ticks to understand the overall diversity of SFG rickettsiae in Japan. Out of 2,189 individuals (19 tick species in 4 genera), 373 (17.0%) samples were positive for Rickettsia spp. as ascertained by real-time PCR amplifcation of the citrate synthase gene (gltA). Conventional PCR and sequencing analyses of gltA indicated the presence of 15 diferent genotypes of SFG rickettsiae. Based on the analysis of fve additional genes, we characterised fve Rickettsia species; R. asiatica, R. helvetica, R. monacensis (formerly reported as Rickettsia sp. In56 in Japan), R. tamurae, and Candidatus R. tarasevichiae and several unclassifed SFG rickettsiae. We also found a strong association between rickettsial genotypes and their host tick species, while there was little association between rickettsial genotypes and their geographical origins.