Detection and Partial Molecular Characterization of Rickettsia and Bartonella from Southern African Bat Species
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bartonella Apis Sp. Nov., a Honey Bee Gut Symbiont of the Class Alphaproteobacteria
Serveur Academique´ Lausannois SERVAL serval.unil.ch Author Manuscript Faculty of Biology and Medicine Publication This paper has been peer-reviewed but does not include the final publisher proof-corrections or journal pagination. Published in final edited form as: Title: Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Authors: Keˇsnerov´aL, Moritz R, Engel P Journal: International journal of systematic and evolutionary microbiology Year: 2016 Jan Issue: 66 Volume: 1 Pages: 414-21 DOI: 10.1099/ijsem.0.000736 In the absence of a copyright statement, users should assume that standard copyright protection applies, unless the article contains an explicit statement to the contrary. In case of doubt, contact the journal publisher to verify the copyright status of an article. 1 Bartonella apis sp. nov., a honey bee gut symbiont of the 2 class Alphaproteobacteria 3 4 Lucie Kešnerová, Roxane Moritz, Philipp Engel* 5 6 Department of Fundamental Microbiology, University of Lausanne, CH-1015 7 Lausanne, Switzerland 8 9 Running title: Description of a bee gut symbiont 10 11 *Correspondence: 12 Prof. Philipp Engel 13 Department of Fundamental Microbiology 14 University of Lausanne, CH-1015 Lausanne, Switzerland 15 Tel.: +41 (0)21 692 56 12 16 e-mail: [email protected] 17 18 Category: New Taxa – Proteobacteria 19 Keywords: Apis mellifera; insect; Bartonella; gut microbiota; Alpha-1 20 21 Sequence deposition: The 16S rRNA gene sequences and protein-coding gene 22 sequences of the bacterial strains PEB0122T, PEB0149, PEB0150, BBC0104, and 23 BBC0108 from Apis mellifera, and the uncultured Rhizobiales bacterium from 24 Herpagnathos saltator are deposited in GenBank with accession numbers KP987849 25 – KP987886 and KT315729 – KT315734. -

Abstract Pultorak, Elizabeth Lauren
ABSTRACT PULTORAK, ELIZABETH LAUREN. The Epidemiology of Lyme Disease and Bartonellosis in Humans and Animals. (Under the direction of Edward B. Breitschwerdt). The expansion of vector borne diseases in humans, a variety of mammalian hosts, and arthropod vectors draws attention to the need for enhanced diagnostic techniques for documenting infection in hosts, effective vector control, and treatment of individuals with associated diseases. Through improved diagnosis of vector-borne disease in both humans and animals, epidemiological studies to elucidate clinical associations or spatio-temporal relationships can be assessed. Veterinarians, through the use of the C6 peptide in the SNAP DX test kit, may be able to evaluate the changing epidemiology of borreliosis through their canine population. We developed a survey to evaluate the practices and perceptions of veterinarians in North Carolina regarding borreliosis in dogs across different geographic regions of the state. We found that veterinarians’ perception of the risk of borreliosis in North Carolina was consistent with recent scientific reports pertaining to geographic expansion of borreliosis in the state. Veterinarians should promote routine screening of dogs for Borrelia burgdorferi exposure as a simple, inexpensive form of surveillance in this transitional geographic region. We next conducted two separate studies to evaluate Bartonella spp. bacteremia or presence of antibodies against B. henselae, B. koehlerae, or B. vinsonii subsp. berkhoffii in 296 patients examined by a rheumatologist and 192 patients with animal exposure (100%) and recent animal bites and scratches (88.0%). Among 296 patients examined by a rheumatologist, prevalence of antibodies (185 [62%]) and Bartonella spp. bacteremia (122 [41.1%]) was high. -

Beating Chronic LYME
Section 1 Beating Chronic LYME Dr. Kevin Conners Fellowship in Integrative Cancer Therapy Fellowship in Anti-Aging, Regenerative, and Functional Medicine American Academy of Anti-Aging Medicine www.ConnersClinic.com From left to right: Larvae, Nymph, Female, Male Tick Tick in Nymph stage is the size of a poppy seed. Beating Chronic LYME Forward I used to live in the woods. My wife and four children at the time purchased 280 acres in Wisconsin and basically lived off the land. We grew most of our own food, were ‘off grid’ as we produced our own electricity through solar panels, and had to pump our water by hand. It was certainly a different way of life that prepared us for missionary work in Mexico. It was 1997 and at the time I had begun hearing about Lyme disease being a tick- born disorder. I had never seen a deer tick before moving to our ‘little house in the big woods’ but one thing was for certain – we had plenty of deer. They were as populous as the mosquitoes. To make a long story short, both my oldest daughter and I had contracted Lyme during our three year stay. I experienced the violent sickness of acute Lyme as well as a beautiful bulls-eye rash that made the diagnosis easy. I took just 3 days of antibiotics and since that was just the second time that I had ever taken a prescription medication in my life, they eradicated the disease effectively. My daughter on the other hand wasn’t so fortunate. She never had an acute illness and we never saw any rash therefore we didn’t catch the disease until it had advanced to Chronic Lyme Disease (CLD). -

Human Case of Bartonella Alsatica Lymphadenitis
LETTERS (6). The sequence is distinct from a DOI: 10.3201/eid1412.080944 Human Case of small number of sequences derived from rabies viruses in Vietnam, which References Bartonella alsatica suggests that China is a stronger can- Lymphadenitis 1. Smith JS, Fishbein DB, Rupprecht CE, didate for the source of the virus than Clark K. Unexplained rabies in three To the Editor: Lymph node en- her native country. immigrants in the United States: a vi- Although the case history could rologic investigation. N Engl J Med. largement is a common medical prob- not provide evidence for interaction 1991;324:205–11. lem that is usually caused by bacterial, 2. Grattan-Smith PJ, O’Regan WJ, Ellis PS, with a dog while her family was in viral, fungal, or protozoal agents (1). O’Flaherty SJ, McIntyre PB, Barnes CJ. A Malignancies or lymphoproliferative Hong Kong Special Administrative second Australian case, with a long incuba- Region, rabies was endemic within tion period. Med J Aust. 1992;156:651–4. diseases are often found, especially in the colony at the time that the pa- 3. McColl KA, Gould AR, Selleck PW, elderly patients (1). Bartonella hense- Hooper PT, Westbury HA, Smith JS. tient’s family was resident. From 1980 lae, the main causative agent of cat- Polymerase chain reaction and other labo- scratch disease (CSD), appears to be through 1984, 5 human cases were re- ratory techniques in the diagnosis of long corded (9). Only 2 case-patients had incubation rabies in Australia. Aust Vet the most common organism respon- clear evidence of a dog bite; histories J. -

Detecting Phylogenetic Signals from Deep Roots of the Tree of Life
UNIVERSITY OF CALIFORNIA,MERCED Detecting Phylogenetic Signals From Deep Roots of the Tree of Life A dissertation submitted in partial fulfillment of the requirements for the degree Doctor of Philosophy in Quantitative and Systems Biology by Katherine Colleen Harris Amrine Committee in charge: Professor Carolin Frank, Chair Professor David Ardell Professor Meng-Lin Tsao Professor Suzanne Sindi August 2013 Copyright Katherine C. Amrine All Rights Reserved UNIVERSITY OF CALIFORNIA,MERCED Graduate Division The Dissertation of Katherine Colleen Harris Amrine is approved, and it is acceptable in quality and form for publication on microfilm and electronically: Faculty Advisor: David H. Ardell Committee Members: Chair: Carolin Frank Meng-Lin Tsao Suzanne Sindi Date iii Contents List of Figures ................................................................. vi List of Tables .................................................................. ix Acknowledgements ............................................................. x Vita ........................................................................... xi Abstract ...................................................................... xii 1 Shifting focus in evolutionary biology – identifying a new signal for phylogenetic tree reconstruction and taxonomic classification 1 1.1 The evolution of bacterial classification and phylogeny . .1 1.2 The historical marker – 16S . .2 1.3 Complications in bacterial classification and phylogeny . .2 1.3.1 Horizontal gene transfer . .2 1.3.2 Does a true tree exist? . .3 1.4 Methods for phylogenetic tree reconstruction . .3 1.4.1 DNA . .3 1.4.2 RNA . .4 1.4.3 Proteins . .4 1.4.4 Data compilation . .5 1.5 Bias in tree-building . .5 1.6 Biological bias in biological data . .6 1.7 The tRNA interaction network . .6 1.8 Information theory . .8 1.9 Machine Learning for bacterial classification . .9 2 tRNA signatures reveal polyphyletic origins of streamlined SAR11 genomes among the Alphaproteobacteria 12 2.1 Abstract . -

Characterizing the Molecular Biology of a Bacteriophage-Like Particle from Bartonella Bacilliformis
University of Montana ScholarWorks at University of Montana Graduate Student Theses, Dissertations, & Professional Papers Graduate School 1999 Characterizing the molecular biology of a bacteriophage-like particle from Bartonella bacilliformis Kent D. Barbian The University of Montana Follow this and additional works at: https://scholarworks.umt.edu/etd Let us know how access to this document benefits ou.y Recommended Citation Barbian, Kent D., "Characterizing the molecular biology of a bacteriophage-like particle from Bartonella bacilliformis" (1999). Graduate Student Theses, Dissertations, & Professional Papers. 6634. https://scholarworks.umt.edu/etd/6634 This Thesis is brought to you for free and open access by the Graduate School at ScholarWorks at University of Montana. It has been accepted for inclusion in Graduate Student Theses, Dissertations, & Professional Papers by an authorized administrator of ScholarWorks at University of Montana. For more information, please contact [email protected]. i I S Maureen and Mike MANSFIELD LIBRARY The University of IVIONTANA Permission is granted by the author to reproduce this material in its entirety, provided that this material is used for scholarly purposes and is properly cited in published works and reports. ** Please check ’’Yes'* or ”No” and provide signature ** Yes, I grant permission X" No, I do not grant permission _____ Author's Signature Date Any copying for conunercial purposes or financial gain may be undertaken only with the author's explicit consent. Characterizing the Molecular Biology of a Bacteriophage-Like Particle FromBartonella bacilliformis by Kent D. Barbian B.A,, The University of Montana, 1997 Presented in partial fulfillment of the requirements for the degree of Master of Science in Microbiology The University of Montana 1999 Approved by: Committee Chair Dean, Graduate School Date UMI Number: EP37435 All rights reserved INFORMATION TO ALL USERS The quality of this reproduction is dependent upon the quality of the copy submitted. -
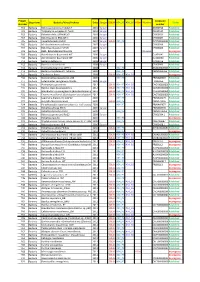
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Viewed to Possess Different Related Sequences Or Outliers and N-Gram-Based Dot Languages
Osmanbeyoglu and Ganapathiraju BMC Bioinformatics 2011, 12:12 http://www.biomedcentral.com/1471-2105/12/12 RESEARCHARTICLE Open Access N-gram analysis of 970 microbial organisms reveals presence of biological language models Hatice Ulku Osmanbeyoglu, Madhavi K Ganapathiraju* Abstract Background: It has been suggested previously that genome and proteome sequences show characteristics typical of natural-language texts such as “signature-style” word usage indicative of authors or topics, and that the algorithms originally developed for natural language processing may therefore be applied to genome sequences to draw biologically relevant conclusions. Following this approach of ‘biological language modeling’, statistical n-gram analysis has been applied for comparative analysis of whole proteome sequences of 44 organisms. It has been shown that a few particular amino acid n-grams are found in abundance in one organism but occurring very rarely in other organisms, thereby serving as genome signatures. At that time proteomes of only 44 organisms were available, thereby limiting the generalization of this hypothesis. Today nearly 1,000 genome sequences and corresponding translated sequences are available, making it feasible to test the existence of biological language models over the evolutionary tree. Results: We studied whole proteome sequences of 970 microbial organisms using n-gram frequencies and cross- perplexity employing the Biological Language Modeling Toolkit and Patternix Revelio toolkit. Genus-specific signatures were observed even in a simple unigram distribution. By taking statistical n-gram model of one organism as reference and computing cross-perplexity of all other microbial proteomes with it, cross-perplexity was found to be predictive of branch distance of the phylogenetic tree. -

Bartonella Spp. Isolated from Wild and Domestic Ruminants in North America1
Dispatches Bartonella spp. Isolated from Wild and Domestic Ruminants in North America1 Chao-chin Chang,* Bruno B. Chomel,* Rickie W. Kasten,* Remy Heller,† Katherine M. Kocan,‡ Hiroshi Ueno,§ Kazuhiro Yamamoto,* Vernon C. Bleich,¶ Becky M. Pierce,¶ Ben J. Gonzales,¶ Pamela K. Swift,¶ Walter M. Boyce,* Spencer S. Jang,* Henri-Jean Boulouis,# and Yves Piémont† *School of Veterinary Medicine, University of California, Davis, California, USA; †Institut de Bactériologie, Université Louis Pasteur, Strasbourg, France; ‡College of Veterinary Medicine, Oklahoma State University, Stillwater, Oklahoma, USA; §School of Veterinary Medicine, Rakuno-Gakuen University, Ebetsu, Hokkaido, Japan; ¶California Department of Fish and Game, Bishop, Rancho Cordova, California, USA; #Ecole Nationale Vétérinaire d’Alfort, 94704 Maisons-Alfort, France Bartonella species were isolated from 49% of 128 cattle from California and Oklahoma, 90% of 42 mule deer from California, and 15% of 100 elk from California and Oregon. Isolates from all 63 cattle, 14 deer, and 1 elk had the same polymerase chain reaction/restriction fragment length polymorphism profiles. Our findings indicate potential for inter- and intraspecies transmission among ruminants, as well as risk that these Bartonella spp. could act as zoonotic agents. Bartonella species have been identified as rRNA and citrate synthase genes (14). Modes of important zoonotic agents (1,2). Cats are the transmission in these ruminants need to be main reservoir of Bartonella henselae, the agent established. Tick transmission has been suspect- that causes cat scratch disease in humans (1). ed but not yet proven for dogs infected with Long-term bacteremia in cats and flea transmis- B. vinsonii subsp. berkhoffii (16). Since fleas are sion from cat to cat, as confirmed by experimental less likely than ticks to infest cattle (17), ticks infection, support a vectorborne transmission (3). -

Human Bartonellosis: an Underappreciated Public Health Problem?
Tropical Medicine and Infectious Disease Review Human Bartonellosis: An Underappreciated Public Health Problem? Mercedes A. Cheslock and Monica E. Embers * Division of Immunology, Tulane National Primate Research Center, Tulane University Health Sciences, Covington, LA 70433, USA; [email protected] * Correspondence: [email protected]; Tel.: +(985)-871-6607 Received: 24 March 2019; Accepted: 16 April 2019; Published: 19 April 2019 Abstract: Bartonella spp. bacteria can be found around the globe and are the causative agents of multiple human diseases. The most well-known infection is called cat-scratch disease, which causes mild lymphadenopathy and fever. As our knowledge of these bacteria grows, new presentations of the disease have been recognized, with serious manifestations. Not only has more severe disease been associated with these bacteria but also Bartonella species have been discovered in a wide range of mammals, and the pathogens’ DNA can be found in multiple vectors. This review will focus on some common mammalian reservoirs as well as the suspected vectors in relation to the disease transmission and prevalence. Understanding the complex interactions between these bacteria, their vectors, and their reservoirs, as well as the breadth of infection by Bartonella around the world will help to assess the impact of Bartonellosis on public health. Keywords: Bartonella; vector; bartonellosis; ticks; fleas; domestic animals; human 1. Introduction Several Bartonella spp. have been linked to emerging and reemerging human diseases (Table1)[ 1–5]. These fastidious, gram-negative bacteria cause the clinically complex disease known as Bartonellosis. Historically, the most common causative agents for human disease have been Bartonella bacilliformis, Bartonella quintana, and Bartonella henselae. -

Ru 2015 150 263 a (51) Мпк A61k 31/155 (2006.01)
РОССИЙСКАЯ ФЕДЕРАЦИЯ (19) (11) (13) RU 2015 150 263 A (51) МПК A61K 31/155 (2006.01) ФЕДЕРАЛЬНАЯ СЛУЖБА ПО ИНТЕЛЛЕКТУАЛЬНОЙ СОБСТВЕННОСТИ (12) ЗАЯВКА НА ИЗОБРЕТЕНИЕ (21)(22) Заявка: 2015150263, 01.05.2014 (71) Заявитель(и): НЕОКУЛИ ПТИ ЛТД (AU) Приоритет(ы): (30) Конвенционный приоритет: (72) Автор(ы): 01.05.2013 AU 2013901517 ПЕЙДЖ Стефен (AU), ГАРГ Санджай (AU) (43) Дата публикации заявки: 06.06.2017 Бюл. № 16 RU (85) Дата начала рассмотрения заявки PCT на национальной фазе: 01.12.2015 (86) Заявка PCT: AU 2014/000480 (01.05.2014) 2015150263 (87) Публикация заявки PCT: WO 2014/176634 (06.11.2014) Адрес для переписки: 190000, Санкт-Петербург, Box-1125, "ПАТЕНТИКА" A (54) СПОСОБЫ ЛЕЧЕНИЯ БАКТЕРИАЛЬНЫХ ИНФЕКЦИЙ (57) Формула изобретения 1. Способ лечения или профилактики бактериальной колонизации или инфекции у субъекта, включающий стадию: введения субъекту терапевтически эффективного количества робенидина или его терапевтически приемлемой соли, причем указанная A бактериальная колонизация или инфекция вызвана бактериальным агентом. 2. Способ по п. 1, отличающийся тем, что субъект выбран из группы, включающей: человека, животных, принадлежащих видам семейства псовых, кошачьих, крупного рогатого скота, овец, коз, свиней, птиц, рыб и лошадей. 3. Способ по п. 1, отличающийся тем, что робенидин вводят субъекту в дозе в диапазоне от 0,1 до 250 мг/кг массы тела. 4. Способ по любому из пп. 1-3, отличающийся тем, что бактериальный агент является 2015150263 грамположительным. 5. Способ по п. 4, отличающийся тем, что бактериальный агент выбран из -

Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales
Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales TECHNICAL BIOSAFETY COMMITTEE (TBC) NATIONAL CENTER FOR GENETIC ENGINEERING AND BIOTECHNOLOGY (BIOTEC) NATIONAL SCIENCE AND TECHNOLOGY DEVELOPMENT AGENCY (NSTDA) MINISTRY OF SCIENCE AND TECHNOLOGY (MOST) 2015 Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales TECHNICAL BIOSAFETY COMMITTEE (TBC) NATIONAL CENTER FOR GENETIC ENGINEERING AND BIOTECHNOLOGY (BIOTEC) NATIONAL SCIENCE AND TECHNOLOGY DEVELOPMENT AGENCY (NSTDA) MINISTRY OF SCIENCE AND TECHNOLOGY (MOST) 2015 Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales Technical Biosafety Committee National Center for Genetic Engineering and Biotechnology National Science and Technology Development Agency (NSTDA) © National Center for Genetic Engineering and Biotechnology 2015 ISBN : 978-616-12-0386-3 Tel : +66(0)2-564-6700 Fax : +66(0)2-564-6703 E-mail : [email protected] URL : http://www.biotec.or.th Printing House : P.A. Living Printing Co.,Ltd 4 Soi Sirintron 7 Road Sirintron District Bangplad Province Bangkok 10700 Tel : +66(0)2-881 9890 Fax : +66(0)2-881 9894 Preface Genetically Modified Microorganisms (GMMs) were first used in B.E. 2525 to produce insulin in industrial medicine. Currently, GMMs are used in various industries, such as the food, pharmaceutical and bioplastic industries, to manufacture a number of important consumer products. To ensure operator and environmental safety, the Technical Biosafety Committee (TBC) of the National Center for Genetic Engineering and Biotechnology (BIOTEC), the National Science and Technology Development Agency (NSTDA), has prepared guidelines for GMM work, publishing “Biosafety Guidelines for Contained Use of Genetically Modified Microorganisms at Pilot and Industrial Scales” in B.E.