Old and New Tick-Borne Rickettsioses
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

Hplc-Uv Quantitation of Folate Synthesized by Rickettsia
HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) By Junyan Chen A Thesis Presented to The Faculty of Humboldt State University In Partial Fulfillment of the Requirements for the Degree Master of Science in Biology Committee Membership Dr. Jianmin Zhong, Committee Chair Dr. David S. Baston, Committee Member Dr. Jenny Cappuccio, Committee Member Dr. Jacob Varkey, Committee Member Dr. Erik Jules, Program Graduate Coordinator December 2017 ABSTRACT HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) Junyan Chen Ticks are the most important vector of many infectious diseases in the United States. Understanding the nature of the relationship between Rickettsia endosymbiont Ixodes pacificus (REIP) and Exudes pacificus will help develop strategies for the control of tick- borne diseases, such as Lyme disease, and Rocky Mountain spotted fever. Folate, also known as vitamin B9, is a necessary vitamin for tick survival, and plays a central role in one-carbon metabolism in cells. Folate exist as a large family of structurally related forms that transfer one-carbon groups among biomolecules that are important to cell growth, differentiation, and survival. In Dr. Zheng’s lab, REIP were cultured in Ixodes scapularis embryonic tick cell line ISE6. Previous research has shown that REIP in Ixodes pacificus carries all five de novo folate biosynthesis genes. Folate biosynthesis mRNAs were detected and all recombinant rickettsial folate proteins were overexpressed. To determine whether REIP synthesize folate, we sought to measure the folate concentration in REIP using HPLC-UV quantification with a Diamond HydrideTM liquid chromatography column. 5-methyltetrahydrofolate (5-MTHF), the active circulating form of folate in bacteria was detected. -

Molecular Evidence of Novel Spotted Fever Group Rickettsia
pathogens Article Molecular Evidence of Novel Spotted Fever Group Rickettsia Species in Amblyomma albolimbatum Ticks from the Shingleback Skink (Tiliqua rugosa) in Southern Western Australia Mythili Tadepalli 1, Gemma Vincent 1, Sze Fui Hii 1, Simon Watharow 2, Stephen Graves 1,3 and John Stenos 1,* 1 Australian Rickettsial Reference Laboratory, University Hospital Geelong, Geelong 3220, Australia; [email protected] (M.T.); [email protected] (G.V.); [email protected] (S.F.H.); [email protected] (S.G.) 2 Reptile Victoria Inc., Melbourne 3035, Australia; [email protected] 3 Department of Microbiology and Infectious Diseases, Nepean Hospital, NSW Health Pathology, Penrith 2747, Australia * Correspondence: [email protected] Abstract: Tick-borne infectious diseases caused by obligate intracellular bacteria of the genus Rick- ettsia are a growing global problem to human and animal health. Surveillance of these pathogens at the wildlife interface is critical to informing public health strategies to limit their impact. In Australia, reptile-associated ticks such as Bothriocroton hydrosauri are the reservoirs for Rickettsia honei, the causative agent of Flinders Island spotted fever. In an effort to gain further insight into the potential for reptile-associated ticks to act as reservoirs for rickettsial infection, Rickettsia-specific PCR screening was performed on 64 Ambylomma albolimbatum ticks taken from shingleback skinks (Tiliqua rugosa) lo- cated in southern Western Australia. PCR screening revealed 92% positivity for rickettsial DNA. PCR Citation: Tadepalli, M.; Vincent, G.; amplification and sequencing of phylogenetically informative rickettsial genes (ompA, ompB, gltA, Hii, S.F.; Watharow, S.; Graves, S.; Stenos, J. -

Babela Massiliensis, a Representative of a Widespread Bacterial
Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier, Natalya Yutin, Olivier Croce, Kira S Makarova, Yuri I Wolf, Samia Benamar, Didier Raoult, Eugene V. Koonin, Bernard La Scola To cite this version: Isabelle Pagnier, Natalya Yutin, Olivier Croce, Kira S Makarova, Yuri I Wolf, et al.. Babela mas- siliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae. Biology Direct, BioMed Central, 2015, 10 (13), 10.1186/s13062-015-0043-z. hal-01217089 HAL Id: hal-01217089 https://hal-amu.archives-ouvertes.fr/hal-01217089 Submitted on 19 Oct 2015 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Pagnier et al. Biology Direct (2015) 10:13 DOI 10.1186/s13062-015-0043-z RESEARCH Open Access Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier1, Natalya Yutin2, Olivier Croce1, Kira S Makarova2, Yuri I Wolf2, Samia Benamar1, Didier Raoult1, Eugene V Koonin2 and Bernard La Scola1* Abstract Background: Only a small fraction of bacteria and archaea that are identifiable by metagenomics can be grown on standard media. -

Diagnostic Code Descriptions (ICD9)
INFECTIONS AND PARASITIC DISEASES INTESTINAL AND INFECTIOUS DISEASES (001 – 009.3) 001 CHOLERA 001.0 DUE TO VIBRIO CHOLERAE 001.1 DUE TO VIBRIO CHOLERAE EL TOR 001.9 UNSPECIFIED 002 TYPHOID AND PARATYPHOID FEVERS 002.0 TYPHOID FEVER 002.1 PARATYPHOID FEVER 'A' 002.2 PARATYPHOID FEVER 'B' 002.3 PARATYPHOID FEVER 'C' 002.9 PARATYPHOID FEVER, UNSPECIFIED 003 OTHER SALMONELLA INFECTIONS 003.0 SALMONELLA GASTROENTERITIS 003.1 SALMONELLA SEPTICAEMIA 003.2 LOCALIZED SALMONELLA INFECTIONS 003.8 OTHER 003.9 UNSPECIFIED 004 SHIGELLOSIS 004.0 SHIGELLA DYSENTERIAE 004.1 SHIGELLA FLEXNERI 004.2 SHIGELLA BOYDII 004.3 SHIGELLA SONNEI 004.8 OTHER 004.9 UNSPECIFIED 005 OTHER FOOD POISONING (BACTERIAL) 005.0 STAPHYLOCOCCAL FOOD POISONING 005.1 BOTULISM 005.2 FOOD POISONING DUE TO CLOSTRIDIUM PERFRINGENS (CL.WELCHII) 005.3 FOOD POISONING DUE TO OTHER CLOSTRIDIA 005.4 FOOD POISONING DUE TO VIBRIO PARAHAEMOLYTICUS 005.8 OTHER BACTERIAL FOOD POISONING 005.9 FOOD POISONING, UNSPECIFIED 006 AMOEBIASIS 006.0 ACUTE AMOEBIC DYSENTERY WITHOUT MENTION OF ABSCESS 006.1 CHRONIC INTESTINAL AMOEBIASIS WITHOUT MENTION OF ABSCESS 006.2 AMOEBIC NONDYSENTERIC COLITIS 006.3 AMOEBIC LIVER ABSCESS 006.4 AMOEBIC LUNG ABSCESS 006.5 AMOEBIC BRAIN ABSCESS 006.6 AMOEBIC SKIN ULCERATION 006.8 AMOEBIC INFECTION OF OTHER SITES 006.9 AMOEBIASIS, UNSPECIFIED 007 OTHER PROTOZOAL INTESTINAL DISEASES 007.0 BALANTIDIASIS 007.1 GIARDIASIS 007.2 COCCIDIOSIS 007.3 INTESTINAL TRICHOMONIASIS 007.8 OTHER PROTOZOAL INTESTINAL DISEASES 007.9 UNSPECIFIED 008 INTESTINAL INFECTIONS DUE TO OTHER ORGANISMS -

Phenotypic and Genomic Analyses of Burkholderia Stabilis Clinical Contamination, Switzerland Helena M.B
RESEARCH Phenotypic and Genomic Analyses of Burkholderia stabilis Clinical Contamination, Switzerland Helena M.B. Seth-Smith, Carlo Casanova, Rami Sommerstein, Dominik M. Meinel,1 Mohamed M.H. Abdelbary,2 Dominique S. Blanc, Sara Droz, Urs Führer, Reto Lienhard, Claudia Lang, Olivier Dubuis, Matthias Schlegel, Andreas Widmer, Peter M. Keller,3 Jonas Marschall, Adrian Egli A recent hospital outbreak related to premoistened gloves pathogens that generally fall within the B. cepacia com- used to wash patients exposed the difficulties of defining plex (Bcc) (1). Burkholderia bacteria have large, flexible, Burkholderia species in clinical settings. The outbreak strain multi-replicon genomes, a large metabolic repertoire, vari- displayed key B. stabilis phenotypes, including the inabil- ous virulence factors, and inherent resistance to many anti- ity to grow at 42°C; we used whole-genome sequencing to microbial drugs (2,3). confirm the pathogen was B. stabilis. The outbreak strain An outbreak of B. stabilis was identified among hos- genome comprises 3 chromosomes and a plasmid, shar- ing an average nucleotide identity of 98.4% with B. stabilis pitalized patients across several cantons in Switzerland ATCC27515 BAA-67, but with 13% novel coding sequenc- during 2015–2016 (4). The bacterium caused bloodstream es. The genome lacks identifiable virulence factors and has infections, noninvasive infections, and wound contamina- no apparent increase in encoded antimicrobial drug resis- tions. The source of the infection was traced to contaminat- tance, few insertion sequences, and few pseudogenes, ed commercially available, premoistened washing gloves suggesting this outbreak was an opportunistic infection by used for bedridden patients. After hospitals discontinued an environmental strain not adapted to human pathogenic- use of these gloves, the outbreak resolved. -

Article/25/5/18-0438-App1.Pdf)
RESEARCH LETTERS Pathology. 2011;43:58–63. http://dx.doi.org/10.1097/ variabilis ticks can transmit the causative agent of Rocky PAT.0b013e328340e431 Mountain spotted fever, and Ixodes scapularis ticks can 8. Rodriguez-Lozano J, Pérez-Llantada E, Agüero J, Rodríguez-Fernández A, Ruiz de Alegria C, Martinez-Martinez L, transmit the causative agents of Lyme disease, babesiosis, et al. Sternal wound infection caused by Gordonia bronchialis: and human granulocytic anaplasmosis (1). Although less identification by MALDI-TOF MS. JMM Case Rep. 2016;3: common in the region, A. maculatum ticks are dominant e005067. in specific habitats and can transmit the causative agent of Rickettsia parkeri rickettsiosis (1). Address for correspondence: Rene Choi, Department of Ophthalmology, Persons who have occupations that require them to be Casey Eye Institute, Oregon Health and Science University, 3375 SW outside on a regular basis might have a greater risk for ac- Terwilliger Blvd, Portland, OR 97239, USA; email: [email protected] quiring a tickborne disease (2). Although numerous stud- ies have been conducted regarding risks for tickborne dis- eases among forestry workers in Europe, few studies have been performed in the United States (2,3). The studies that have been conducted in the United States have focused on forestry workers in the northeastern region (2). However, because of variable phenology and densities of ticks, it is useful to evaluate tick activity and pathogen prevalence in Rickettsiales in Ticks various regions and ecosystems. Burn-tolerant and burn-dependent ecosystems, such as Removed from Outdoor pine (Pinus spp.) and mixed pine forests commonly found Workers, Southwest Georgia in the southeastern United States, have unique tick dynam- and Northwest Florida, USA ics compared with those of other habitats (4). -

Case Report: Coinfection with Rickettsia Monacensis and Orientia Tsutsugamushi
Am. J. Trop. Med. Hyg., 101(2), 2019, pp. 332–335 doi:10.4269/ajtmh.18-0631 Copyright © 2019 by The American Society of Tropical Medicine and Hygiene Case Report: Coinfection with Rickettsia monacensis and Orientia tsutsugamushi Seok Won Kim,1† Choon-Mee Kim,2† Dong-Min Kim,3* and Na Ra Yun3 1Department of Neurosurgery, College of Medicine, Chosun University, Gwangju, Republic of Korea; 2Premedical Science, College of Medicine, Chosun University, Gwangju, Republic of Korea; 3Department of Internal Medicine, College of Medicine, Chosun University, Gwangju, Republic of Korea Abstract. Rickettsia monacensis and Orientia tsutsugamushi are bacteria of the family Rickettsiaceae, which causes fever, rash, and eschar formation; outdoor activities are a risk factor for Rickettsiaceae infection. A 75-year-old woman presented with fever, rash, and eschar and was confirmed as being scrub typhus based on a nested-polymerase chain reaction (N-PCR) test for a 56-kDa gene of O. tsutsugamushi; the genome was identified as the Boryong genotype. In addition, a pan-Rickettsia real-time PCR test was positive and a N-PCR test using a Rickettsia-specific partial outer membrane protein A (rOmpA) confirmed R. monacensis. This is the first case wherein a patient suspected of having scrub typhus owing to the presence of rash and eschar was also found to be coinfected with O. tsutsugamushi and R. monacensis based on molecular testing. INTRODUCTION leukocyte count, 7,200/mm3; hemoglobin, 11.6 g/dL; platelet count, 232,000/mm3; and erythrocyte sedimentation rate, 31 Rickettsia monacensis is a pathogen that causes spotted mm/hours. C-reactive protein and procalcitonin levels were fever group rickettsial infection; the main symptoms of in- elevated at 9.26 mg/dL and 0.836 ng/mL (0–0.5 ng/mL), re- fection include fever, headache, and myalgia, as well as es- 1 spectively. -
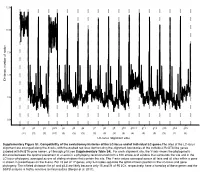
LC-Locus Alignment Sites Distance, Number of Nodes Supplementary
12.5 10.0 7.5 5.0 Distance, number of nodes 2.5 0.0 g1 g2 g3 g3.5 g4 g5 g6 g7 g8 g9 g10 g10.1 g11 g12 g13 g14 g15 (11) (3) (3) (10) (3) (3) (3) (3) (3) (3) (3) (4) (3) (3) (3) (2) (3) LC-locus alignment sites Supplementary Figure S1. Compatibility of the evolutionary histories of the LC-locus and of individual LC genes.The sites of the LC-locus alignment are arranged along the X-axis, with the dashed red lines demarcating the alignment boundaries of the individual RcGTA-like genes (labeled with RcGTA gene names, g1 through g15; see Supplementary Table S4). For each alignment site, the Y-axis shows the phylogenetic distance between the optimal placement of a taxon in a phylogeny reconstructed from a 100 amino-acid window that surrounds the site and in the LC-locus phylogeny, averaged across all sliding windows that contain the site. The Y-axis values averaged across all taxa and all sites within a gene is shown in parentheses on the X-axis. For 15 out of 17 genes, only 2-4 nodes separate the optimal taxon position in the LC-locus and gene phylogeny. The inflated distances for g1 and g3.5 are likely because only 15 and 21 of 95 LCs, respectively, have a homolog of these genes and the SSPB analysis is highly sensitive to missing data (Berger et al. 2011). a. Bacteria Unassigned Thermotogae Tenericutes Synergistetes Spirochaetes Proteobacteria ylum Planctomycetes h p Firmicutes Deferribacteres Cyanobacteria Chloroflexi Bacteroidetes Actinobacteria Acidobacteria 1(11,750) 2(1,750) 3(2,538) 4(168) 5(51) 6(54) 7(43) 8(32) 9(26) 10(40) 11(33) 12(198) 13(173) 14(101) 15(98) 16(43) 17(114) Number of rcc01682−rcc01698 homologs in a cluster b. -

Genome Project Reveals a Putative Rickettsial Endosymbiont
GBE Bacterial DNA Sifted from the Trichoplax adhaerens (Animalia: Placozoa) Genome Project Reveals a Putative Rickettsial Endosymbiont Timothy Driscoll1,y, Joseph J. Gillespie1,2,*,y, Eric K. Nordberg1,AbduF.Azad2, and Bruno W. Sobral1,3 1Virginia Bioinformatics Institute at Virginia Polytechnic Institute and State University 2Department of Microbiology and Immunology, University of Maryland School of Medicine 3Present address: Nestle´ Institute of Health Sciences SA, Campus EPFL, Quartier de L’innovation, Lausanne, Switzerland *Corresponding author: E-mail: [email protected]. yThese authors contributed equally to this work. Accepted: March 1, 2013 Abstract Eukaryotic genome sequencing projects often yield bacterial DNA sequences, data typically considered as microbial contamination. However, these sequences may also indicate either symbiont genes or lateral gene transfer (LGT) to host genomes. These bacterial sequences can provide clues about eukaryote–microbe interactions. Here, we used the genome of the primitive animal Trichoplax adhaerens (Metazoa: Placozoa), which is known to harbor an uncharacterized Gram-negative endosymbiont, to search for the presence of bacterial DNA sequences. Bioinformatic and phylogenomic analyses of extracted data from the genome assembly (181 bacterial coding sequences [CDS]) and trace read archive (16S rDNA) revealed a dominant proteobacterial profile strongly skewed to Rickettsiales (Alphaproteobacteria) genomes. By way of phylogenetic analysis of 16S rDNA and 113 proteins conserved across proteobacterial genomes, as well as identification of 27 rickettsial signature genes, we propose a Rickettsiales endosymbiont of T. adhaerens (RETA). The majority (93%) of the identified bacterial CDS belongs to small scaffolds containing prokaryotic-like genes; however, 12 CDS were identified on large scaffolds comprised of eukaryotic-like genes, suggesting that T. -

Rickettsialpox-A Newly Recognized Rickettsial Disease V
Public Health Reports Vol. 62 * MAY 30, 1947 * No. 22 Printed With the Approval of the Bureau of the Budget as Required by Rule 42 of the Joint-Committee on Printing RICKETTSIALPOX-A NEWLY RECOGNIZED RICKETTSIAL DISEASE V. RECOVERY OF RICKETTSIA AKARI FROM A HOUSE MOUSE (MUS MUSCULUS)1 By ROBERT J. HUEBNER, Senior Assistant Surgeon, WILLIAm L. JELLISON, Parasitologist, CHARLES ARMSTRONG, Medical Director, United States Public Health Service Ricketttia akari, the causative agent of rickettsialpox, was isolated from the blood of persons ill with this disease (1) and from rodent mites Allodermanyssus sanguineus Hirst inhabiting the domicile of ill per- sons (2). This paper describes the isolation of R. akari from a house mouse (Mus musculus) trapped on the same premises-a housing development in the citr of New York where more than 100 cases of rickettsialpox have occurred (3), (4), (5), (6). Approximately 60 house mice were trapped in the basements of this housing development where rodent harborage existed in store rooms and in incinerator ashpits. Engorged mites were occasionally found attached to the mice, the usual site of attachment being the rump. Mites were frequently found inside the box traps after the captured mice were removed. Early attempts to isolate the etiological agent of rickettisalpox from these mice were complicated by the presence of choriomeningitis among them. Twelve successive suspensions of mouse tissue, repre- senting 16 house mice, inoculated intracerebrally into laboratory mice (Swiss strain) and intraperitoneally into guinea pigs resulted in the production of a highly lethal disease in both species which was identified immunologically as choriomeningitis. -

Ohio Department of Health, Bureau of Infectious Diseases Disease Name Class A, Requires Immediate Phone Call to Local Health
Ohio Department of Health, Bureau of Infectious Diseases Reporting specifics for select diseases reportable by ELR Class A, requires immediate phone Susceptibilities specimen type Reportable test name (can change if Disease Name other specifics+ call to local health required* specifics~ state/federal case definition or department reporting requirements change) Culture independent diagnostic tests' (CIDT), like BioFire panel or BD MAX, E. histolytica Stain specimen = stool, bile results should be sent as E. histolytica DNA fluid, duodenal fluid, 260373001^DETECTED^SCT with E. histolytica Antigen Amebiasis (Entamoeba histolytica) No No tissue large intestine, disease/organism-specific DNA LOINC E. histolytica Antibody tissue small intestine codes OR a generic CIDT-LOINC code E. histolytica IgM with organism-specific DNA SNOMED E. histolytica IgG codes E. histolytica Total Antibody Ova and Parasite Anthrax Antibody Anthrax Antigen Anthrax EITB Acute Anthrax EITB Convalescent Anthrax Yes No Culture ELISA PCR Stain/microscopy Stain/spore ID Eastern Equine Encephalitis virus Antibody Eastern Equine Encephalitis virus IgG Antibody Eastern Equine Encephalitis virus IgM Arboviral neuroinvasive and non- Eastern Equine Encephalitis virus RNA neuroinvasive disease: Eastern equine California serogroup virus Antibody encephalitis virus disease; LaCrosse Equivocal results are accepted for all California serogroup virus IgG Antibody virus disease (other California arborviral diseases; California serogroup virus IgM Antibody specimen = blood, serum, serogroup -

Rickettsia Helvetica in Dermacentor Reticulatus Ticks
DISPATCHES The Study Rickettsia helvetica Using the cloth-dragging method, during March–May 2007 we collected 100 adult Dermacentor spp. ticks from in Dermacentor meadows in 2 different locations near Cakovec, between the Drava and Mura rivers in the central part of Medjimurje Coun- reticulatus Ticks ty. This area is situated in the northwestern part of Croatia, at Marinko Dobec, Dragutin Golubic, 46″38′N, 16″43′E, and has a continental climate with an Volga Punda-Polic, Franz Kaeppeli, average annual air temperature of 10.4°C at an altitude of and Martin Sievers 164 m. To isolate DNA from ticks, we modifi ed the method We report on the molecular evidence that Dermacentor used by Nilsson et al. (11). Before DNA isolation, ticks reticulatus ticks in Croatia are infected with Rickettsia hel- were disinfected in 70% ethanol and dried. Each tick was vetica (10%) or Rickettsia slovaca (2%) or co-infected with mechanically crushed in a Dispomix 25 tube with lysis buf- both species (1%). These fi ndings expand the knowledge of fer by using the Dispomix (Medic Tools, Zug, Switzerland). the geographic distribution of R. helvetica and D. reticulatus Lysis of each of the crushed tick samples was carried out in ticks. a solution of 6.7% sucrose, 0.2% proteinase K, 20 mg/mL lysozyme, and 10 ng/ml RNase A for 16 h at 37°C; 0.5 mo- ickettsia helvetica organisms were fi rst isolated from lar EDTA, and 20% sodium dodecyl sulfate was added and RIxodes ricinus ticks in Switzerland and were consid- further incubated for 1 h at 37°C.