POLMIT Partenaire.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Hplc-Uv Quantitation of Folate Synthesized by Rickettsia
HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) By Junyan Chen A Thesis Presented to The Faculty of Humboldt State University In Partial Fulfillment of the Requirements for the Degree Master of Science in Biology Committee Membership Dr. Jianmin Zhong, Committee Chair Dr. David S. Baston, Committee Member Dr. Jenny Cappuccio, Committee Member Dr. Jacob Varkey, Committee Member Dr. Erik Jules, Program Graduate Coordinator December 2017 ABSTRACT HPLC-UV QUANTITATION OF FOLATE SYNTHESIZED BY RICKETTSIA ENDOSYMBIONT IXODES PACIFICUS (REIP) Junyan Chen Ticks are the most important vector of many infectious diseases in the United States. Understanding the nature of the relationship between Rickettsia endosymbiont Ixodes pacificus (REIP) and Exudes pacificus will help develop strategies for the control of tick- borne diseases, such as Lyme disease, and Rocky Mountain spotted fever. Folate, also known as vitamin B9, is a necessary vitamin for tick survival, and plays a central role in one-carbon metabolism in cells. Folate exist as a large family of structurally related forms that transfer one-carbon groups among biomolecules that are important to cell growth, differentiation, and survival. In Dr. Zheng’s lab, REIP were cultured in Ixodes scapularis embryonic tick cell line ISE6. Previous research has shown that REIP in Ixodes pacificus carries all five de novo folate biosynthesis genes. Folate biosynthesis mRNAs were detected and all recombinant rickettsial folate proteins were overexpressed. To determine whether REIP synthesize folate, we sought to measure the folate concentration in REIP using HPLC-UV quantification with a Diamond HydrideTM liquid chromatography column. 5-methyltetrahydrofolate (5-MTHF), the active circulating form of folate in bacteria was detected. -

Metaproteogenomic Insights Beyond Bacterial Response to Naphthalene
ORIGINAL ARTICLE ISME Journal – Original article Metaproteogenomic insights beyond bacterial response to 5 naphthalene exposure and bio-stimulation María-Eugenia Guazzaroni, Florian-Alexander Herbst, Iván Lores, Javier Tamames, Ana Isabel Peláez, Nieves López-Cortés, María Alcaide, Mercedes V. del Pozo, José María Vieites, Martin von Bergen, José Luis R. Gallego, Rafael Bargiela, Arantxa López-López, Dietmar H. Pieper, Ramón Rosselló-Móra, Jesús Sánchez, Jana Seifert and Manuel Ferrer 10 Supporting Online Material includes Text (Supporting Materials and Methods) Tables S1 to S9 Figures S1 to S7 1 SUPPORTING TEXT Supporting Materials and Methods Soil characterisation Soil pH was measured in a suspension of soil and water (1:2.5) with a glass electrode, and 5 electrical conductivity was measured in the same extract (diluted 1:5). Primary soil characteristics were determined using standard techniques, such as dichromate oxidation (organic matter content), the Kjeldahl method (nitrogen content), the Olsen method (phosphorus content) and a Bernard calcimeter (carbonate content). The Bouyoucos Densimetry method was used to establish textural data. Exchangeable cations (Ca, Mg, K and 10 Na) extracted with 1 M NH 4Cl and exchangeable aluminium extracted with 1 M KCl were determined using atomic absorption/emission spectrophotometry with an AA200 PerkinElmer analyser. The effective cation exchange capacity (ECEC) was calculated as the sum of the values of the last two measurements (sum of the exchangeable cations and the exchangeable Al). Analyses were performed immediately after sampling. 15 Hydrocarbon analysis Extraction (5 g of sample N and Nbs) was performed with dichloromethane:acetone (1:1) using a Soxtherm extraction apparatus (Gerhardt GmbH & Co. -

E. Coli (Expec) Among E
Elucidating the Unknown Ecology of Bacterial Pathogens from Genomic Data Tristan Kishan Seecharran A thesis submitted in partial fulfilment of the requirements of Nottingham Trent University for the degree of Doctor of Philosophy June 2018 Copyright Statement I hereby declare that the work presented in this thesis is the result of original research carried out by the author, unless otherwise stated. No material contained herein has been submitted for any other degree, or at any other institution. This work is an intellectual property of the author. You may copy up to 5% of this work for private study, or personal, non-commercial research. Any re-use of the information contained within this document should be fully referenced, quoting the author, title, university, degree level and pagination. Queries or requests for any other use, or if a more substantial copy is required, should be directed in the owner(s) of the Intellectual Property Rights. Tristan Kishan Seecharran i Acknowledgements I would like to express my sincere gratitude and thanks to my external advisor Alan McNally and director of studies Ben Dickins for their continued support, guidance and encouragement, and without whom, the completion of this thesis would not have been possible. Many thanks also go to the members of the Pathogen Research Group at Nottingham Trent University. I would like to thank Gina Manning and Jody Winter in particular for their invaluable advice and contributions during lab meetings. I would also like to thank our collaborators, Mikael Skurnik and colleagues from the University of Helsinki and Jukka Corander from the University of Oslo, for their much-appreciated support and assistance in this project and the published work on Yersinia pseudotuberculosis. -

Evolutionary Origin of Insect–Wolbachia Nutritional Mutualism
Evolutionary origin of insect–Wolbachia nutritional mutualism Naruo Nikoha,1, Takahiro Hosokawab,1, Minoru Moriyamab,1, Kenshiro Oshimac, Masahira Hattoric, and Takema Fukatsub,2 aDepartment of Liberal Arts, The Open University of Japan, Chiba 261-8586, Japan; bBioproduction Research Institute, National Institute of Advanced Industrial Science and Technology, Tsukuba 305-8566, Japan; and cCenter for Omics and Bioinformatics, Graduate School of Frontier Sciences, University of Tokyo, Kashiwa 277-8561, Japan Edited by Nancy A. Moran, University of Texas at Austin, Austin, TX, and approved June 3, 2014 (received for review May 20, 2014) Obligate insect–bacterium nutritional mutualism is among the insects, generally conferring negative fitness consequences to most sophisticated forms of symbiosis, wherein the host and the their hosts and often causing hosts’ reproductive aberrations to symbiont are integrated into a coherent biological entity and un- enhance their own transmission in a selfish manner (7, 8). Re- able to survive without the partnership. Originally, however, such cently, however, a Wolbachia strain associated with the bedbug obligate symbiotic bacteria must have been derived from free-living Cimex lectularius,designatedaswCle, was shown to be es- bacteria. How highly specialized obligate mutualisms have arisen sential for normal growth and reproduction of the blood- from less specialized associations is of interest. Here we address this sucking insect host via provisioning of B vitamins (9). Hence, it –Wolbachia evolutionary -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

Genome Project Reveals a Putative Rickettsial Endosymbiont
GBE Bacterial DNA Sifted from the Trichoplax adhaerens (Animalia: Placozoa) Genome Project Reveals a Putative Rickettsial Endosymbiont Timothy Driscoll1,y, Joseph J. Gillespie1,2,*,y, Eric K. Nordberg1,AbduF.Azad2, and Bruno W. Sobral1,3 1Virginia Bioinformatics Institute at Virginia Polytechnic Institute and State University 2Department of Microbiology and Immunology, University of Maryland School of Medicine 3Present address: Nestle´ Institute of Health Sciences SA, Campus EPFL, Quartier de L’innovation, Lausanne, Switzerland *Corresponding author: E-mail: [email protected]. yThese authors contributed equally to this work. Accepted: March 1, 2013 Abstract Eukaryotic genome sequencing projects often yield bacterial DNA sequences, data typically considered as microbial contamination. However, these sequences may also indicate either symbiont genes or lateral gene transfer (LGT) to host genomes. These bacterial sequences can provide clues about eukaryote–microbe interactions. Here, we used the genome of the primitive animal Trichoplax adhaerens (Metazoa: Placozoa), which is known to harbor an uncharacterized Gram-negative endosymbiont, to search for the presence of bacterial DNA sequences. Bioinformatic and phylogenomic analyses of extracted data from the genome assembly (181 bacterial coding sequences [CDS]) and trace read archive (16S rDNA) revealed a dominant proteobacterial profile strongly skewed to Rickettsiales (Alphaproteobacteria) genomes. By way of phylogenetic analysis of 16S rDNA and 113 proteins conserved across proteobacterial genomes, as well as identification of 27 rickettsial signature genes, we propose a Rickettsiales endosymbiont of T. adhaerens (RETA). The majority (93%) of the identified bacterial CDS belongs to small scaffolds containing prokaryotic-like genes; however, 12 CDS were identified on large scaffolds comprised of eukaryotic-like genes, suggesting that T. -

Ohio Department of Health, Bureau of Infectious Diseases Disease Name Class A, Requires Immediate Phone Call to Local Health
Ohio Department of Health, Bureau of Infectious Diseases Reporting specifics for select diseases reportable by ELR Class A, requires immediate phone Susceptibilities specimen type Reportable test name (can change if Disease Name other specifics+ call to local health required* specifics~ state/federal case definition or department reporting requirements change) Culture independent diagnostic tests' (CIDT), like BioFire panel or BD MAX, E. histolytica Stain specimen = stool, bile results should be sent as E. histolytica DNA fluid, duodenal fluid, 260373001^DETECTED^SCT with E. histolytica Antigen Amebiasis (Entamoeba histolytica) No No tissue large intestine, disease/organism-specific DNA LOINC E. histolytica Antibody tissue small intestine codes OR a generic CIDT-LOINC code E. histolytica IgM with organism-specific DNA SNOMED E. histolytica IgG codes E. histolytica Total Antibody Ova and Parasite Anthrax Antibody Anthrax Antigen Anthrax EITB Acute Anthrax EITB Convalescent Anthrax Yes No Culture ELISA PCR Stain/microscopy Stain/spore ID Eastern Equine Encephalitis virus Antibody Eastern Equine Encephalitis virus IgG Antibody Eastern Equine Encephalitis virus IgM Arboviral neuroinvasive and non- Eastern Equine Encephalitis virus RNA neuroinvasive disease: Eastern equine California serogroup virus Antibody encephalitis virus disease; LaCrosse Equivocal results are accepted for all California serogroup virus IgG Antibody virus disease (other California arborviral diseases; California serogroup virus IgM Antibody specimen = blood, serum, serogroup -

Gene Gain and Loss Events in Rickettsia and Orientia Species Kalliopi Georgiades1,2, Vicky Merhej1, Khalid El Karkouri1, Didier Raoult1, Pierre Pontarotti2*
Georgiades et al. Biology Direct 2011, 6:6 http://www.biology-direct.com/content/6/1/6 RESEARCH Open Access Gene gain and loss events in Rickettsia and Orientia species Kalliopi Georgiades1,2, Vicky Merhej1, Khalid El Karkouri1, Didier Raoult1, Pierre Pontarotti2* Abstract Background: Genome degradation is an ongoing process in all members of the Rickettsiales order, which makes these bacterial species an excellent model for studying reductive evolution through interspecies variation in genome size and gene content. In this study, we evaluated the degree to which gene loss shaped the content of some Rickettsiales genomes. We shed light on the role played by horizontal gene transfers in the genome evolution of Rickettsiales. Results: Our phylogenomic tree, based on whole-genome content, presented a topology distinct from that of the whole core gene concatenated phylogenetic tree, suggesting that the gene repertoires involved have different evolutionary histories. Indeed, we present evidence for 3 possible horizontal gene transfer events from various organisms to Orientia and 6 to Rickettsia spp., while we also identified 3 possible horizontal gene transfer events from Rickettsia and Orientia to other bacteria. We found 17 putative genes in Rickettsia spp. that are probably the result of de novo gene creation; 2 of these genes appear to be functional. On the basis of these results, we were able to reconstruct the gene repertoires of “proto-Rickettsiales” and “proto-Rickettsiaceae”, which correspond to the ancestors of Rickettsiales and Rickettsiaceae, respectively. Finally, we found that 2,135 genes were lost during the evolution of the Rickettsiaceae to an intracellular lifestyle. Conclusions: Our phylogenetic analysis allowed us to track the gene gain and loss events occurring in bacterial genomes during their evolution from a free-living to an intracellular lifestyle. -

Table S5. the Information of the Bacteria Annotated in the Soil Community at Species Level
Table S5. The information of the bacteria annotated in the soil community at species level No. Phylum Class Order Family Genus Species The number of contigs Abundance(%) 1 Firmicutes Bacilli Bacillales Bacillaceae Bacillus Bacillus cereus 1749 5.145782459 2 Bacteroidetes Cytophagia Cytophagales Hymenobacteraceae Hymenobacter Hymenobacter sedentarius 1538 4.52499338 3 Gemmatimonadetes Gemmatimonadetes Gemmatimonadales Gemmatimonadaceae Gemmatirosa Gemmatirosa kalamazoonesis 1020 3.000970902 4 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas indica 797 2.344876284 5 Firmicutes Bacilli Lactobacillales Streptococcaceae Lactococcus Lactococcus piscium 542 1.594633558 6 Actinobacteria Thermoleophilia Solirubrobacterales Conexibacteraceae Conexibacter Conexibacter woesei 471 1.385742446 7 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas taxi 430 1.265115184 8 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas wittichii 388 1.141545794 9 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas sp. FARSPH 298 0.876754244 10 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sorangium cellulosum 260 0.764953367 11 Proteobacteria Deltaproteobacteria Myxococcales Polyangiaceae Sorangium Sphingomonas sp. Cra20 260 0.764953367 12 Proteobacteria Alphaproteobacteria Sphingomonadales Sphingomonadaceae Sphingomonas Sphingomonas panacis 252 0.741416341 -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -
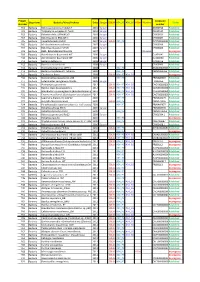
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Epidemiology and Comparative Analysis of Yersinia in Ireland Author(S) Ringwood, Tamara Publication Date 2013 Original Citation Ringwood, T
UCC Library and UCC researchers have made this item openly available. Please let us know how this has helped you. Thanks! Title Epidemiology and comparative analysis of Yersinia in Ireland Author(s) Ringwood, Tamara Publication date 2013 Original citation Ringwood, T. 2013. Epidemiology and comparative analysis of Yersinia in Ireland. PhD Thesis, University College Cork. Type of publication Doctoral thesis Rights © 2013, Tamara Ringwood http://creativecommons.org/licenses/by-nc-nd/3.0/ Item downloaded http://hdl.handle.net/10468/1294 from Downloaded on 2021-10-07T12:07:10Z Epidemiology and Comparative Analysis of Yersinia in Ireland by Tamara Ringwood A thesis presented for the Degree of Doctor of Philosophy National University of Ireland, Cork University College Cork Coláiste na hOllscoile Corcaigh Department of Microbiology Head of Department: Prof. Gerald F. Fitzgerald Supervisor: Prof. Michael B. Prentice April 2013 Contents List of Tables .............................................................................................................................................. iv List of figures ............................................................................................................................................. vi Declaration ............................................................................................................................................... viii Acknowledgements ................................................................................................................................