18167638.Pdf
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metaproteogenomic Insights Beyond Bacterial Response to Naphthalene
ORIGINAL ARTICLE ISME Journal – Original article Metaproteogenomic insights beyond bacterial response to 5 naphthalene exposure and bio-stimulation María-Eugenia Guazzaroni, Florian-Alexander Herbst, Iván Lores, Javier Tamames, Ana Isabel Peláez, Nieves López-Cortés, María Alcaide, Mercedes V. del Pozo, José María Vieites, Martin von Bergen, José Luis R. Gallego, Rafael Bargiela, Arantxa López-López, Dietmar H. Pieper, Ramón Rosselló-Móra, Jesús Sánchez, Jana Seifert and Manuel Ferrer 10 Supporting Online Material includes Text (Supporting Materials and Methods) Tables S1 to S9 Figures S1 to S7 1 SUPPORTING TEXT Supporting Materials and Methods Soil characterisation Soil pH was measured in a suspension of soil and water (1:2.5) with a glass electrode, and 5 electrical conductivity was measured in the same extract (diluted 1:5). Primary soil characteristics were determined using standard techniques, such as dichromate oxidation (organic matter content), the Kjeldahl method (nitrogen content), the Olsen method (phosphorus content) and a Bernard calcimeter (carbonate content). The Bouyoucos Densimetry method was used to establish textural data. Exchangeable cations (Ca, Mg, K and 10 Na) extracted with 1 M NH 4Cl and exchangeable aluminium extracted with 1 M KCl were determined using atomic absorption/emission spectrophotometry with an AA200 PerkinElmer analyser. The effective cation exchange capacity (ECEC) was calculated as the sum of the values of the last two measurements (sum of the exchangeable cations and the exchangeable Al). Analyses were performed immediately after sampling. 15 Hydrocarbon analysis Extraction (5 g of sample N and Nbs) was performed with dichloromethane:acetone (1:1) using a Soxtherm extraction apparatus (Gerhardt GmbH & Co. -

Evolutionary Origin of Insect–Wolbachia Nutritional Mutualism
Evolutionary origin of insect–Wolbachia nutritional mutualism Naruo Nikoha,1, Takahiro Hosokawab,1, Minoru Moriyamab,1, Kenshiro Oshimac, Masahira Hattoric, and Takema Fukatsub,2 aDepartment of Liberal Arts, The Open University of Japan, Chiba 261-8586, Japan; bBioproduction Research Institute, National Institute of Advanced Industrial Science and Technology, Tsukuba 305-8566, Japan; and cCenter for Omics and Bioinformatics, Graduate School of Frontier Sciences, University of Tokyo, Kashiwa 277-8561, Japan Edited by Nancy A. Moran, University of Texas at Austin, Austin, TX, and approved June 3, 2014 (received for review May 20, 2014) Obligate insect–bacterium nutritional mutualism is among the insects, generally conferring negative fitness consequences to most sophisticated forms of symbiosis, wherein the host and the their hosts and often causing hosts’ reproductive aberrations to symbiont are integrated into a coherent biological entity and un- enhance their own transmission in a selfish manner (7, 8). Re- able to survive without the partnership. Originally, however, such cently, however, a Wolbachia strain associated with the bedbug obligate symbiotic bacteria must have been derived from free-living Cimex lectularius,designatedaswCle, was shown to be es- bacteria. How highly specialized obligate mutualisms have arisen sential for normal growth and reproduction of the blood- from less specialized associations is of interest. Here we address this sucking insect host via provisioning of B vitamins (9). Hence, it –Wolbachia evolutionary -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -
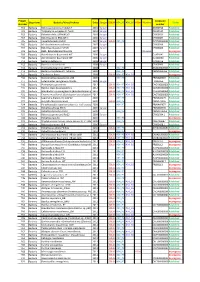
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Potential Role of Bacteria Packaging by Protozoa in the Persistence and Transmission of Pathogenic Bacteria
HYPOTHESIS AND THEORY ARTICLE published: 21 May 2014 doi: 10.3389/fmicb.2014.00240 Potential role of bacteria packaging by protozoa in the persistence and transmission of pathogenic bacteria Alix M. Denoncourt 1,2, Valérie E. Paquet 1,2 and Steve J. Charette 1,2,3* 1 Institut de Biologie Intégrative et des Systèmes, Université Laval, Quebec City, QC, Canada 2 Centre de Recherche de l’Institut Universitaire de Cardiologie et de Pneumologie de Québec, Quebec City, QC, Canada 3 Département de Biochimie, de Microbiologie et de Bio-Informatique, Faculté des Sciences et de Génie, Université Laval, Quebec City, QC, Canada Edited by: Many pathogenic bacteria live in close association with protozoa. These unicellular Mickael Desvaux, INRA, France eukaryotic microorganisms are ubiquitous in various environments. A number of protozoa Reviewed by: such as amoebae and ciliates ingest pathogenic bacteria, package them usually in O Roger Anderson, Lamont-Doherty membrane structures, and then release them into the environment. Packaged bacteria Earth Observatory of Columbia University, USA are more resistant to various stresses and are more apt to survive than free bacteria. Stephan Schmitz-Esser, University New evidence indicates that protozoa and not bacteria control the packaging process. It of Veterinary Medicine Vienna, is possible that packaging is more common than suspected and may play a major role in Austria the persistence and transmission of pathogenic bacteria. To confirm the role of packaging Thomas Hindré, University J. Fourier, France in the propagation of infections, it is vital that the molecular mechanisms governing the *Correspondence: packaging of bacteria by protozoa be identified as well as elements related to the ecology Steve J. -

Supporting Information
Supporting Information McLean et al. “Candidate Phylum TM6 Genome Recovered from a Hospital Sink Biofilm Provides the First Genomic Insights into this Uncultivated Phylum “ SI Appendix McLean et al. PNAS Supporting Information a b High Fluor TM6 Background 1 Fig. S1. Biofilm sample biomass and Fluorescence Activated Cell Sorting (FACS) plot of a 2 biofilm sample. a) Sample biomass collected directly into buffer solution and from a biofilm in 3 a sink drain within a restroom adjacent to an emergency waiting room. b) Sorting gates set to 4 sort events after staining with SYBR Green DNA stain. The P1 gate includes high fluorescent 5 SYBR Green stained particles, and the background gate indicates that region in which unstained 6 sample events were located. The low fluorescent P2 region was chosen as a sort gate to target a 7 total of 100 events in each of 32 wells of a 384 well plate. 1 Fig. S2. Custom integrated Agilent Technologies BioCel 1200 liquid handling automated 2 platform for high throughput single cell genomics. The BioCel platform allows processing of 3 more than 5,000 single cells per week through a multi-stage protocol that includes multiple 4 displacement amplification (MDA) of DNA, MDA dilution and 16S PCR, MDA and PCR hit 5 picking, Picogreen (Life Technologies) DNA quantitation, 16S Syto 9 (Life Technologies) melt 6 curve assay, 16S Taqman qPCR, and SAP/Exonuclease I (Affymetrix) PCR treatment. All liquid 7 handling is performed on the BioCel with the BioRAPTR (Beckman Coulter) and Bravo 8 (Agilent) performing non-contact dispensing and liquid transfer steps, respectively. -

Babela Massiliensis, a Representative of A
Pagnier et al. Biology Direct (2015) 10:13 DOI 10.1186/s13062-015-0043-z RESEARCH Open Access Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae Isabelle Pagnier1, Natalya Yutin2, Olivier Croce1, Kira S Makarova2, Yuri I Wolf2, Samia Benamar1, Didier Raoult1, Eugene V Koonin2 and Bernard La Scola1* Abstract Background: Only a small fraction of bacteria and archaea that are identifiable by metagenomics can be grown on standard media. Recent efforts on deep metagenomics sequencing, single-cell genomics and the use of specialized culture conditions (culturomics) increasingly yield novel microbes some of which represent previously uncharacterized phyla and possess unusual biological traits. Results: We report isolation and genome analysis of Babela massiliensis, an obligate intracellular parasite of Acanthamoeba castellanii. B. massiliensis shows an unusual, fission mode of cell multiplication whereby large, polymorphic bodies accumulate in the cytoplasm of infected amoeba and then split into mature bacterial cells. This unique mechanism of cell division is associated with a deep degradation of the cell division machinery and delayed expression of the ftsZ gene. The genome of B. massiliensis consists of a circular chromosome approximately 1.12 megabase in size that encodes, 981 predicted proteins, 38 tRNAs and one typical rRNA operon. Phylogenetic analysis shows that B. massiliensis belongs to the putative bacterial phylum TM6 that so far was represented by the draft genome of the JCVI TM6SC1 bacterium obtained by single cell genomics and numerous environmental sequences. Conclusions: Currently, B. massiliensis is the only cultivated member of the putative TM6 phylum. Phylogenomic analysis shows diverse taxonomic affinities for B. -

Downloaded from the Virulence Factor of Pathogenic Bacteria Database (VFDB)70
www.nature.com/scientificreports OPEN Investigation of potential pathogenicity of Willaertia magna by investigating the transfer of bacteria pathogenicity genes into its genome Issam Hasni1,2, Nisrine Chelkha1, Emeline Baptiste1, Mouh Rayane Mameri2, Joel Lachuer3,4,5, Fabrice Plasson2, Philippe Colson 1 & Bernard La Scola1* Willaertia magna c2c maky is a thermophilic amoeba closely related to the genus Naegleria. This free-living amoeba has the ability to eliminate Legionella pneumophila, which is an amoeba-resisting bacterium living in an aquatic environment. To prevent the proliferation of L. pneumophila in cooling towers, the use of W. magna as natural biocide has been proposed. To provide a better understanding of the W. magna genome, whole-genome sequencing was performed through the study of virulence factors and lateral gene transfers. This amoeba harbors a genome of 36.5 megabases with 18,519 predicted genes. BLASTp analyses reported protein homology between 136 W. magna sequences and amoeba-resistant microorganisms. Horizontal gene transfers were observed based on the basis of the phylogenetic reconstruction hypothesis. We detected 15 homologs of N. fowleri genes related to virulence, although these latter were also found in the genome of N. gruberi, which is a non-pathogenic amoeba. Furthermore, the cytotoxicity test performed on human cells supports the hypothesis that the strain c2c maky is a non-pathogenic amoeba. This work explores the genomic repertory for the frst draft genome of genus Willaertia and provides genomic data for further comparative studies on virulence of related pathogenic amoeba, N. fowleri. Heterolobosea is a rich class of species of heterotrophs, almost all free-living, containing pathogenic (e.g. -

Author Manuscript Faculty of Biology and Medicine Publication
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Serveur académique lausannois Serveur Académique Lausannois SERVAL serval.unil.ch Author Manuscript Faculty of Biology and Medicine Publication This paper has been peer-reviewed but dos not include the final publisher proof-corrections or journal pagination. Published in final edited form as: Title: Importance of amoebae as a tool to isolate amoeba-resisting microorganisms and for their ecology and evolution: the Chlamydia paradigm. Authors: Kebbi-Beghdadi C, Greub G Journal: Environmental microbiology reports Year: 2014 Aug Volume: 6 Issue: 4 Pages: 309-24 DOI: 10.1111/1758-2229.12155 In the absence of a copyright statement, users should assume that standard copyright protection applies, unless the article contains an explicit statement to the contrary. In case of doubt, contact the journal publisher to verify the copyright status of an article. 1 1 Importance of amoebae as a tool to isolate amoeba-resisting microorganisms and 2 for their ecology and evolution: the Chlamydia paradigm. 3 4 Carole Kebbi-Beghdadi and Gilbert Greub 5 Center for Research on Intracellular Bacteria (CRIB), Institute of Microbiology, University 6 Hospital Center and University of Lausanne, Lausanne, Switzerland. 7 8 9 10 11 12 13 Corresponding author: 14 Dr Gilbert Greub 15 Institute of Microbiology 16 Rue du Bugnon 48 17 1011 Lausanne 18 Switzerland 19 Tel: 0041 21 314 4979 20 Fax 0041 21 314 4060 21 e-mail: [email protected] 22 Running title :Discovering new pathogens 23 24 25 26 27 2 1 Summary 2 Free-living amoebae are worldwide distributed and frequently in contact with humans and 3 animals. -

Detection of Legionella Pneumophila in Water and Biofilm Samples by Culture and Molecular Methods from Man-Made Systems in São Paulo - Brazil
Brazilian Journal of Microbiology (2007) 38:743-751 ISSN 1517-8382 DETECTION OF LEGIONELLA PNEUMOPHILA IN WATER AND BIOFILM SAMPLES BY CULTURE AND MOLECULAR METHODS FROM MAN-MADE SYSTEMS IN SÃO PAULO - BRAZIL Fábio R. S. Carvalho; Annette S. Foronda; Vivian H. Pellizari* Laboratório de Microbiologia Ambiental, Departamento de Microbiologia, Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, SP, Brasil. Submitted: January 09, 2007; Returned to authors for corrections: March 30, 2007; Approved: September 20, 2007. ABSTRACT Legionella pneumophila is a pathogenic bacteria associated to aquatic habitat of natural and artificial environments. Clinical cases of legionellosis have been reported in Brazil but there is a lack of information about the incidence and concentration of this bacterium in environmental sources. Thus, the present study was designed to evaluate the occurrence of legionellae in São Paulo city, Brazil, using different methods of detection and identification. Sixty-seven water and biofilm samples from natural reservoirs and man-made systems were collected and analyzed for the presence of Legionella spp by culturing onto a selective medium, coculture in axenic free-living amoebae and direct fluorescent antibody (DFA) assay. Results showed that freshwater of reservoirs did not contain legionellae, Legionella pneumophila was isolated from man-made systems, with predominance of Legionella pneumophila serogroup 1 strains. Although there was no statistical difference among the proposed detection methods, the plate culture method yielded a higher number of L. pneumophila positive samples, followed by amoebic coculture procedure and direct fluorescent antibody assay. Results of PCR and sequencing reactions revealed that application of macrophage infectivity potentiator gene as a molecular marker was an important tool for the identification of environmental isolates of L. -

Exploitation of Host Polyubiquitination Machinery Through Molecular Mimicry by Eukaryotic-Like Bacterial F-Box Effectors
REVIEW ARTICLE published: 11 November 2010 doi: 10.3389/fmicb.2010.00122 Exploitation of host polyubiquitination machinery through molecular mimicry by eukaryotic-like bacterial F-box effectors Christopher T. D. Price1 and Yousef Abu Kwaik1,2,3,4* 1 Department of Microbiology and Immunology, College of Medicine, University of Louisville, Louisville, KY, USA 2 Department of Biology, University of Louisville, Louisville, KY, USA 3 Department of Microbiology, Al-Quds University Medical School, Abu Dies, Jerusalem, Palestinian Territory 4 Department of Microbiology and Molecular Genetics, The Hebrew University Medical School, Jerusalem, Israel Edited by: Microbial pathogens have evolved exquisite mechanisms to interfere and intercept host biological Amal Amer, The Ohio State University, processes, often through molecular mimicry of specific host proteins. Ubiquitination is a highly USA conserved eukaryotic post-translational modification essential in determining protein fate, and Reviewed by: Amal Amer, The Ohio State University, is often hijacked by pathogenic bacteria. The conserved SKP1/CUL1/F-box (SCF) E3 ubiquitin USA ligase complex plays a key role in ubiquitination of proteins in eukaryotic cells. The F-box protein John S. Gunn, The Ohio State component of the SCF complex provides specificity to ubiquitination by binding to specific University, USA cellular proteins, targeting them to be ubiquitinated by the SCF complex. The bacterial pathogens Dario S. Zamboni, Universidade de São Paulo, Brazil Legionella pneumophila, Agrobacterium tumefaciens, and Ralstonia solanacearum utilize type III *Correspondence: or IV translocation systems to inject into the host cell eukaryotic-like F-box effectors that interact Yousef Abu Kwaik, Department of with the host SKP1 component of the SCF complex to trigger ubiquitination of specific host Microbiology and Immunology, College cells targets, which is essential to promote proliferation of these pathogens.