Author Manuscript Faculty of Biology and Medicine Publication
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Metaproteogenomic Insights Beyond Bacterial Response to Naphthalene
ORIGINAL ARTICLE ISME Journal – Original article Metaproteogenomic insights beyond bacterial response to 5 naphthalene exposure and bio-stimulation María-Eugenia Guazzaroni, Florian-Alexander Herbst, Iván Lores, Javier Tamames, Ana Isabel Peláez, Nieves López-Cortés, María Alcaide, Mercedes V. del Pozo, José María Vieites, Martin von Bergen, José Luis R. Gallego, Rafael Bargiela, Arantxa López-López, Dietmar H. Pieper, Ramón Rosselló-Móra, Jesús Sánchez, Jana Seifert and Manuel Ferrer 10 Supporting Online Material includes Text (Supporting Materials and Methods) Tables S1 to S9 Figures S1 to S7 1 SUPPORTING TEXT Supporting Materials and Methods Soil characterisation Soil pH was measured in a suspension of soil and water (1:2.5) with a glass electrode, and 5 electrical conductivity was measured in the same extract (diluted 1:5). Primary soil characteristics were determined using standard techniques, such as dichromate oxidation (organic matter content), the Kjeldahl method (nitrogen content), the Olsen method (phosphorus content) and a Bernard calcimeter (carbonate content). The Bouyoucos Densimetry method was used to establish textural data. Exchangeable cations (Ca, Mg, K and 10 Na) extracted with 1 M NH 4Cl and exchangeable aluminium extracted with 1 M KCl were determined using atomic absorption/emission spectrophotometry with an AA200 PerkinElmer analyser. The effective cation exchange capacity (ECEC) was calculated as the sum of the values of the last two measurements (sum of the exchangeable cations and the exchangeable Al). Analyses were performed immediately after sampling. 15 Hydrocarbon analysis Extraction (5 g of sample N and Nbs) was performed with dichloromethane:acetone (1:1) using a Soxtherm extraction apparatus (Gerhardt GmbH & Co. -

Ptolemeba N. Gen., a Novel Genus of Hartmannellid Amoebae (Tubulinea, Amoebozoa); with an Emphasis on the Taxonomy of Saccamoeba
The Journal of Published by the International Society of Eukaryotic Microbiology Protistologists Journal of Eukaryotic Microbiology ISSN 1066-5234 ORIGINAL ARTICLE Ptolemeba n. gen., a Novel Genus of Hartmannellid Amoebae (Tubulinea, Amoebozoa); with an Emphasis on the Taxonomy of Saccamoeba Pamela M. Watsona, Stephanie C. Sorrella & Matthew W. Browna,b a Department of Biological Sciences, Mississippi State University, Mississippi State, Mississippi, 39762 b Institute for Genomics, Biocomputing & Biotechnology, Mississippi State University, Mississippi State, Mississippi, 39762 Keywords ABSTRACT 18S rRNA; amoeba; amoeboid; Cashia; cristae; freshwater amoebae; Hartmannella; Hartmannellid amoebae are an unnatural assemblage of amoeboid organisms mitochondrial morphology; SSU rDNA; SSU that are morphologically difficult to discern from one another. In molecular phy- rRNA; terrestrial amoebae; tubulinid. logenetic trees of the nuclear-encoded small subunit rDNA, they occupy at least five lineages within Tubulinea, a well-supported clade in Amoebozoa. The Correspondence polyphyletic nature of the hartmannellids has led to many taxonomic problems, M.W. Brown, Department of Biological in particular paraphyletic genera. Recent taxonomic revisions have alleviated Sciences, Mississippi State University, some of the problems. However, the genus Saccamoeba is paraphyletic and is Mississippi State, MS 39762, USA still in need of revision as it currently occupies two distinct lineages. Here, we Telephone number: +1 662-325-2406; report a new clade on the tree of Tubulinea, which we infer represents a novel FAX number: +1 662-325-7939; genus that we name Ptolemeba n. gen. This genus subsumes a clade of hart- e-mail: [email protected] mannellid amoebae that were previously considered in the genus Saccamoeba, but whose mitochondrial morphology is distinct from Saccamoeba. -

Evolutionary Origin of Insect–Wolbachia Nutritional Mutualism
Evolutionary origin of insect–Wolbachia nutritional mutualism Naruo Nikoha,1, Takahiro Hosokawab,1, Minoru Moriyamab,1, Kenshiro Oshimac, Masahira Hattoric, and Takema Fukatsub,2 aDepartment of Liberal Arts, The Open University of Japan, Chiba 261-8586, Japan; bBioproduction Research Institute, National Institute of Advanced Industrial Science and Technology, Tsukuba 305-8566, Japan; and cCenter for Omics and Bioinformatics, Graduate School of Frontier Sciences, University of Tokyo, Kashiwa 277-8561, Japan Edited by Nancy A. Moran, University of Texas at Austin, Austin, TX, and approved June 3, 2014 (received for review May 20, 2014) Obligate insect–bacterium nutritional mutualism is among the insects, generally conferring negative fitness consequences to most sophisticated forms of symbiosis, wherein the host and the their hosts and often causing hosts’ reproductive aberrations to symbiont are integrated into a coherent biological entity and un- enhance their own transmission in a selfish manner (7, 8). Re- able to survive without the partnership. Originally, however, such cently, however, a Wolbachia strain associated with the bedbug obligate symbiotic bacteria must have been derived from free-living Cimex lectularius,designatedaswCle, was shown to be es- bacteria. How highly specialized obligate mutualisms have arisen sential for normal growth and reproduction of the blood- from less specialized associations is of interest. Here we address this sucking insect host via provisioning of B vitamins (9). Hence, it –Wolbachia evolutionary -

Old Woman Creek National Estuarine Research Reserve Management Plan 2011-2016
Old Woman Creek National Estuarine Research Reserve Management Plan 2011-2016 April 1981 Revised, May 1982 2nd revision, April 1983 3rd revision, December 1999 4th revision, May 2011 Prepared for U.S. Department of Commerce Ohio Department of Natural Resources National Oceanic and Atmospheric Administration Division of Wildlife Office of Ocean and Coastal Resource Management 2045 Morse Road, Bldg. G Estuarine Reserves Division Columbus, Ohio 1305 East West Highway 43229-6693 Silver Spring, MD 20910 This management plan has been developed in accordance with NOAA regulations, including all provisions for public involvement. It is consistent with the congressional intent of Section 315 of the Coastal Zone Management Act of 1972, as amended, and the provisions of the Ohio Coastal Management Program. OWC NERR Management Plan, 2011 - 2016 Acknowledgements This management plan was prepared by the staff and Advisory Council of the Old Woman Creek National Estuarine Research Reserve (OWC NERR), in collaboration with the Ohio Department of Natural Resources-Division of Wildlife. Participants in the planning process included: Manager, Frank Lopez; Research Coordinator, Dr. David Klarer; Coastal Training Program Coordinator, Heather Elmer; Education Coordinator, Ann Keefe; Education Specialist Phoebe Van Zoest; and Office Assistant, Gloria Pasterak. Other Reserve staff including Dick Boyer and Marje Bernhardt contributed their expertise to numerous planning meetings. The Reserve is grateful for the input and recommendations provided by members of the Old Woman Creek NERR Advisory Council. The Reserve is appreciative of the review, guidance, and council of Division of Wildlife Executive Administrator Dave Scott and the mapping expertise of Keith Lott and the late Steve Barry. -

New Data on the Cyst Structure of Hartmannella Vermiformis Page, 1967 (Lobosea, Gymnamoebia)
Protistology 1 (2), 82-85 (1999) Protistology July, 1999 New data on the cyst structure of Hartmannella vermiformis Page, 1967 (Lobosea, Gymnamoebia) Alexey V. Smirnov a and Rolf Michel b a Dept. of Invert ebrate Zoology, Fac. of Biology & Soil Sci., St. Petersburg State University, Russia; b Zentrales Institut des Sanitatsdienstes der Bundeswehr Koblenz, Laborabteilung I - Medizin Mikrobiologie, Germany Summary Isolates of widely distributed amoeba species - Hartmannella vermiformis differs in the details of cyst structure. This is unusual for gymnamoebae and is important in course of modern approaches to amoebae systematic. Cysts of the isolate of this species, described here, have regular separation of the cyst wall into endocyst and ectocyst, in contrast with the “original” isolates, described by F.C. Page. Fine structure of H. vermiformis cysts is studied and discussed. Key words: amoebae, Lobosea, Gymnamoebia, Hartmannella, cyst, ultrastructure Introduction portable water-treatment plant near Bonn (Germany). Clonal cultures were maintained on NN-agar plates (Page, Hartmannella vermiformis was described by Page 1988) provided with Enterobacter cloacae as nutrient bac- (1967), and re-investigated twice (Page, 1974, 1985). Tro- teria. All measurements and photographs were made from phozoites of H. vermiformis are very characteristic, and alive amoebae moving on glass surface. identification of this species does not seem to be a prob- For TEM amoebae were fixed for 1h in 3% glutaral- lem. However, cyst structure in this species varies from dehyde with cacodylate buffer (pH 7.2), washed twice in strain to strain. Cyst are double-walled, but ectocyst and the same buffer, postfixed for 1h in 1% osmium tetroxide, endocyst may be so closely apposed in some strains, that dehydrated in ethanol series, and embedded in Spurr resin. -

Legionella Shows a Diverse Secondary Metabolism Dependent on a Broad Spectrum Sfp-Type Phosphopantetheinyl Transferase
Legionella shows a diverse secondary metabolism dependent on a broad spectrum Sfp-type phosphopantetheinyl transferase Nicholas J. Tobias1, Tilman Ahrendt1, Ursula Schell2, Melissa Miltenberger1, Hubert Hilbi2,3 and Helge B. Bode1,4 1 Fachbereich Biowissenschaften, Merck Stiftungsprofessur fu¨r Molekulare Biotechnologie, Goethe Universita¨t, Frankfurt am Main, Germany 2 Max von Pettenkofer Institute, Ludwig-Maximilians-Universita¨tMu¨nchen, Munich, Germany 3 Institute of Medical Microbiology, University of Zu¨rich, Zu¨rich, Switzerland 4 Buchmann Institute for Molecular Life Sciences, Goethe Universita¨t, Frankfurt am Main, Germany ABSTRACT Several members of the genus Legionella cause Legionnaires’ disease, a potentially debilitating form of pneumonia. Studies frequently focus on the abundant number of virulence factors present in this genus. However, what is often overlooked is the role of secondary metabolites from Legionella. Following whole genome sequencing, we assembled and annotated the Legionella parisiensis DSM 19216 genome. Together with 14 other members of the Legionella, we performed comparative genomics and analysed the secondary metabolite potential of each strain. We found that Legionella contains a huge variety of biosynthetic gene clusters (BGCs) that are potentially making a significant number of novel natural products with undefined function. Surprisingly, only a single Sfp-like phosphopantetheinyl transferase is found in all Legionella strains analyzed that might be responsible for the activation of all carrier proteins in primary (fatty acid biosynthesis) and secondary metabolism (polyketide and non-ribosomal peptide synthesis). Using conserved active site motifs, we predict Submitted 29 June 2016 some novel compounds that are probably involved in cell-cell communication, Accepted 25 October 2016 Published 24 November 2016 differing to known communication systems. -

A Revised Classification of Naked Lobose Amoebae (Amoebozoa
Protist, Vol. 162, 545–570, October 2011 http://www.elsevier.de/protis Published online date 28 July 2011 PROTIST NEWS A Revised Classification of Naked Lobose Amoebae (Amoebozoa: Lobosa) Introduction together constitute the amoebozoan subphy- lum Lobosa, which never have cilia or flagella, Molecular evidence and an associated reevaluation whereas Variosea (as here revised) together with of morphology have recently considerably revised Mycetozoa and Archamoebea are now grouped our views on relationships among the higher-level as the subphylum Conosa, whose constituent groups of amoebae. First of all, establishing the lineages either have cilia or flagella or have lost phylum Amoebozoa grouped all lobose amoe- them secondarily (Cavalier-Smith 1998, 2009). boid protists, whether naked or testate, aerobic Figure 1 is a schematic tree showing amoebozoan or anaerobic, with the Mycetozoa and Archamoe- relationships deduced from both morphology and bea (Cavalier-Smith 1998), and separated them DNA sequences. from both the heterolobosean amoebae (Page and The first attempt to construct a congruent molec- Blanton 1985), now belonging in the phylum Per- ular and morphological system of Amoebozoa by colozoa - Cavalier-Smith and Nikolaev (2008), and Cavalier-Smith et al. (2004) was limited by the the filose amoebae that belong in other phyla lack of molecular data for many amoeboid taxa, (notably Cercozoa: Bass et al. 2009a; Howe et al. which were therefore classified solely on morpho- 2011). logical evidence. Smirnov et al. (2005) suggested The phylum Amoebozoa consists of naked and another system for naked lobose amoebae only; testate lobose amoebae (e.g. Amoeba, Vannella, this left taxa with no molecular data incertae sedis, Hartmannella, Acanthamoeba, Arcella, Difflugia), which limited its utility. -

The Risk to Human Health from Free-Living Amoebae Interaction with Legionella in Drinking and Recycled Water Systems
THE RISK TO HUMAN HEALTH FROM FREE-LIVING AMOEBAE INTERACTION WITH LEGIONELLA IN DRINKING AND RECYCLED WATER SYSTEMS Dissertation submitted by JACQUELINE MARIE THOMAS BACHELOR OF SCIENCE (HONOURS) AND BACHELOR OF ARTS, UNSW In partial fulfillment of the requirements for the award of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL ENGINEERING SCHOOL OF CIVIL AND ENVIRONMENTAL ENGINEERING FACULTY OF ENGINEERING MAY 2012 SUPERVISORS Professor Nicholas Ashbolt Office of Research and Development United States Environmental Protection Agency Cincinnati, Ohio USA and School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Professor Richard Stuetz School of Civil and Environmental Engineering Faculty of Engineering The University of New South Wales Sydney, Australia Doctor Torsten Thomas School of Biotechnology and Biomolecular Sciences Faculty of Science The University of New South Wales Sydney, Australia ORIGINALITY STATEMENT '1 hereby declare that this submission is my own work and to the best of my knowledge it contains no materials previously published or written by another person, or substantial proportions of material which have been accepted for the award of any other degree or diploma at UNSW or any other educational institution, except where due acknowledgement is made in the thesis. Any contribution made to the research by others, with whom 1 have worked at UNSW or elsewhere, is explicitly acknowledged in the thesis. I also declare that the intellectual content of this thesis is the product of my own work, except to the extent that assistance from others in the project's design and conception or in style, presentation and linguistic expression is acknowledged.' Signed ~ ............................ -

18167638.Pdf
REVIEW ARTICLE Amoebal pathogens as emerging causal agents of pneumonia Fred´ eric´ Lamoth1 & Gilbert Greub1,2 1Infectious Diseases Service, University of Lausanne, Lausanne, Switzerland; and 2Center for Research on Intracellular Bacteria, Institute of Microbiology, University Hospital Center and University of Lausanne, Lausanne, Switzerland Correspondence: Gilbert Greub, Center for Abstract Research on Intracellular Bacteria, Institute of Microbiology, University Hospital Center and Despite using modern microbiological diagnostic approaches, the aetiological University of Lausanne, Rue du Bugnon 46, agents of pneumonia remain unidentified in about 50% of cases. Some bacteria 1011 Lausanne, Switzerland. Tel.: 141 21 that grow poorly or not at all in axenic media used in routine clinical bacteriology 31449 79; fax: 141 21 31440 60; e-mail: laboratory but which can develop inside amoebae may be the agents of these lower [email protected] respiratory tract infections (RTIs) of unexplained aetiology. Such amoebae- resisting bacteria, which coevolved with amoebae to resist their microbicidal Received 24 September 2009; revised 30 machinery, may have developed virulence traits that help them survive within November 2009; accepted 2 December 2009. human macrophages, i.e. the first line of innate immune defence in the lung. We Final version published online 22 January 2010. review here the current evidence for the emerging pathogenic role of various DOI:10.1111/j.1574-6976.2009.00207.x amoebae-resisting microorganisms as agents of RTIs in humans. Specifically, we discuss the emerging pathogenic roles of Legionella-like amoebal pathogens, novel Editor: Colin Berry Chlamydiae (Parachlamydia acanthamoebae, Simkania negevensis), waterborne mycobacteria and Bradyrhizobiaceae (Bosea and Afipia spp.). Keywords free-living amoebae; amoebae-resisting bacteria; Legionella; Chlamydia-like bacteria; mycobacteria; pneumonia. -

The Microbial Food Web of the Coastal Southern Baltic Sea As Influenced by Wind-Induced Sediment Resuspension
THE MICROBIAL FOOD WEB OF THE COASTAL SOUTHERN BALTIC SEA AS INFLUENCED BY WIND-INDUCED SEDIMENT RESUSPENSION I n a u g u r a l - D i s s e r t a t i o n zur Erlangung des Doktorgrades der Mathematisch-Naturwissenschaftlichen Fakultät der Universität zu Köln vorgelegt von TOBIAS GARSTECKI aus Magdeburg Köln, 2001 Berichterstatter: Prof. Dr. STEPHEN A. WICKHAM Prof. Dr. HARTMUT ARNDT Tag der letzten mündlichen Prüfung: 29.06.2001 Inhaltsverzeichnis Angabe von verwendeten Fremddaten .............................................. 5 Abkürzungsverzeichnis ..................................................................... 6 1. Einleitung ........................................................................................................ 7 1.1. Begründung der Fragestellung ......................................................... 7 1.2. Herangehensweise ..................................................................... 11 2. A comparison of benthic and planktonic heterotrophic protistan community structure in shallow inlets of the Southern Baltic Sea ........… 13 2.1. Summary ................... ........................................................................ 13 2.2. Introduction ................................................................................. 14 2.3. Materials and Methods ..................................................................... 15 2.3.1. Study sites ..................................................................... 15 2.3.2. Sampling design ........................................................ -
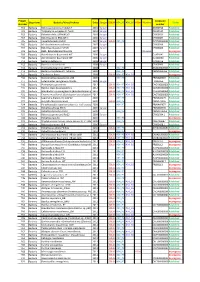
Project Number Organisms Bacteria/Virus/Archaea Date
Project_ Accession Organisms Bacteria/Virus/Archaea Date Sanger SOLiD 454_PE 454_SG PGM Illumina Status Number number P01 Bacteria Rickettsia conorii str.Malish 7 2001 Sanger AE006914 Published P02 Bacteria Tropheryma whipplei str.Twist 2003 Sanger AE014184 Published P03 Bacteria Rickettsia felis URRWXCal2 2005 Sanger CP000053 Published P04 Bacteria Rickettsia bellii RML369-C 2006 Sanger CP000087 Published P05 Bacteria Coxiella burnetii CB109 2007 Sanger SOLiD 454_PE AKYP00000000 Published P06 Bacteria Minibacterium massiliensis 2007 Sanger CP000269 Published P07 Bacteria Rickettsia massiliae MTU5 2007 Sanger CP000683 Published P08 Bacteria BaBL=Bête à Bernard Lascola 2007 Illumina In progress P09 Bacteria Acinetobacter baumannii AYE 2006 Sanger CU459141 Published P10 Bacteria Acinetobacter baumannii SDF 2006 Sanger CU468230 Published P11 Bacteria Borrelia duttonii Ly 2008 Sanger CP000976 Published P12 Bacteria Borrelia recurrentis A1 2008 Sanger CP000993 Published P13 Bacteria Francisella tularensis URFT1 2008 454_PE ABAZ00000000Published P14 Bacteria Borrelia crocidurae str. Achema 2009 454_PE PRJNA162335 Published P15 Bacteria Citrobacter koseri 2009 SOLiD 454_PE 454_SG In progress P16 Bacteria Diplorickettsia massiliensis 20B 2009 454_PE PRJNA86907 Published P17 Bacteria Enterobacter aerogenes EA1509E 2009 Sanger FO203355 Published P18 Bacteria Actinomyces grossensis 2012 SOLiD 454_PE 454_SG CAGY00000000Published P19 Bacteria Bacillus massiliosenegalensis 2012 SOLiD 454_PE 454_SG CAHJ00000000 Published P20 Bacteria Brevibacterium senegalensis -

Lists of Names of Prokaryotic Candidatus Taxa
NOTIFICATION LIST: CANDIDATUS LIST NO. 1 Oren et al., Int. J. Syst. Evol. Microbiol. DOI 10.1099/ijsem.0.003789 Lists of names of prokaryotic Candidatus taxa Aharon Oren1,*, George M. Garrity2,3, Charles T. Parker3, Maria Chuvochina4 and Martha E. Trujillo5 Abstract We here present annotated lists of names of Candidatus taxa of prokaryotes with ranks between subspecies and class, pro- posed between the mid- 1990s, when the provisional status of Candidatus taxa was first established, and the end of 2018. Where necessary, corrected names are proposed that comply with the current provisions of the International Code of Nomenclature of Prokaryotes and its Orthography appendix. These lists, as well as updated lists of newly published names of Candidatus taxa with additions and corrections to the current lists to be published periodically in the International Journal of Systematic and Evo- lutionary Microbiology, may serve as the basis for the valid publication of the Candidatus names if and when the current propos- als to expand the type material for naming of prokaryotes to also include gene sequences of yet-uncultivated taxa is accepted by the International Committee on Systematics of Prokaryotes. Introduction of the category called Candidatus was first pro- morphology, basis of assignment as Candidatus, habitat, posed by Murray and Schleifer in 1994 [1]. The provisional metabolism and more. However, no such lists have yet been status Candidatus was intended for putative taxa of any rank published in the journal. that could not be described in sufficient details to warrant Currently, the nomenclature of Candidatus taxa is not covered establishment of a novel taxon, usually because of the absence by the rules of the Prokaryotic Code.